Brain Landmark Detection
Papers and Code
Benchmarking ERP Analysis: Manual Features, Deep Learning, and Foundation Models
Jan 02, 2026Event-related potential (ERP), a specialized paradigm of electroencephalographic (EEG), reflects neurological responses to external stimuli or events, generally associated with the brain's processing of specific cognitive tasks. ERP plays a critical role in cognitive analysis, the detection of neurological diseases, and the assessment of psychological states. Recent years have seen substantial advances in deep learning-based methods for spontaneous EEG and other non-time-locked task-related EEG signals. However, their effectiveness on ERP data remains underexplored, and many existing ERP studies still rely heavily on manually extracted features. In this paper, we conduct a comprehensive benchmark study that systematically compares traditional manual features (followed by a linear classifier), deep learning models, and pre-trained EEG foundation models for ERP analysis. We establish a unified data preprocessing and training pipeline and evaluate these approaches on two representative tasks, ERP stimulus classification and ERP-based brain disease detection, across 12 publicly available datasets. Furthermore, we investigate various patch-embedding strategies within advanced Transformer architectures to identify embedding designs that better suit ERP data. Our study provides a landmark framework to guide method selection and tailored model design for future ERP analysis. The code is available at https://github.com/DL4mHealth/ERP-Benchmark.
ArteryX: Advancing Brain Artery Feature Extraction with Vessel-Fused Networks and a Robust Validation Framework
Jul 10, 2025Cerebrovascular pathology significantly contributes to cognitive decline and neurological disorders, underscoring the need for advanced tools to assess vascular integrity. Three-dimensional Time-of-Flight Magnetic Resonance Angiography (3D TOF MRA) is widely used to visualize cerebral vasculature, however, clinical evaluations generally focus on major arterial abnormalities, overlooking quantitative metrics critical for understanding subtle vascular changes. Existing methods for extracting structural, geometrical and morphological arterial features from MRA - whether manual or automated - face challenges including user-dependent variability, steep learning curves, and lack of standardized quantitative validations. We propose a novel semi-supervised artery evaluation framework, named ArteryX, a MATLAB-based toolbox that quantifies vascular features with high accuracy and efficiency, achieving processing times ~10-15 minutes per subject at 0.5 mm resolution with minimal user intervention. ArteryX employs a vessel-fused network based landmarking approach to reliably track and manage tracings, effectively addressing the issue of dangling/disconnected vessels. Validation on human subjects with cerebral small vessel disease demonstrated its improved sensitivity to subtle vascular changes and better performance than an existing semi-automated method. Importantly, the ArteryX toolbox enables quantitative feature validation by integrating an in-vivo like artery simulation framework utilizing vessel-fused graph nodes and predefined ground-truth features for specific artery types. Thus, the ArteryX framework holds promise for benchmarking feature extraction toolboxes and for seamless integration into clinical workflows, enabling early detection of cerebrovascular pathology and standardized comparisons across patient cohorts to advance understanding of vascular contributions to brain health.
nnLandmark: A Self-Configuring Method for 3D Medical Landmark Detection
Apr 10, 2025Landmark detection plays a crucial role in medical imaging tasks that rely on precise spatial localization, including specific applications in diagnosis, treatment planning, image registration, and surgical navigation. However, manual annotation is labor-intensive and requires expert knowledge. While deep learning shows promise in automating this task, progress is hindered by limited public datasets, inconsistent benchmarks, and non-standardized baselines, restricting reproducibility, fair comparisons, and model generalizability. This work introduces nnLandmark, a self-configuring deep learning framework for 3D medical landmark detection, adapting nnU-Net to perform heatmap-based regression. By leveraging nnU-Net's automated configuration, nnLandmark eliminates the need for manual parameter tuning, offering out-of-the-box usability. It achieves state-of-the-art accuracy across two public datasets, with a mean radial error (MRE) of 1.5 mm on the Mandibular Molar Landmark (MML) dental CT dataset and 1.2 mm for anatomical fiducials on a brain MRI dataset (AFIDs), where nnLandmark aligns with the inter-rater variability of 1.5 mm. With its strong generalization, reproducibility, and ease of deployment, nnLandmark establishes a reliable baseline for 3D landmark detection, supporting research in anatomical localization and clinical workflows that depend on precise landmark identification. The code will be available soon.
Spatial regularisation for improved accuracy and interpretability in keypoint-based registration
Mar 07, 2025Unsupervised registration strategies bypass requirements in ground truth transforms or segmentations by optimising similarity metrics between fixed and moved volumes. Among these methods, a recent subclass of approaches based on unsupervised keypoint detection stand out as very promising for interpretability. Specifically, these methods train a network to predict feature maps for fixed and moving images, from which explainable centres of mass are computed to obtain point clouds, that are then aligned in closed-form. However, the features returned by the network often yield spatially diffuse patterns that are hard to interpret, thus undermining the purpose of keypoint-based registration. Here, we propose a three-fold loss to regularise the spatial distribution of the features. First, we use the KL divergence to model features as point spread functions that we interpret as probabilistic keypoints. Then, we sharpen the spatial distributions of these features to increase the precision of the detected landmarks. Finally, we introduce a new repulsive loss across keypoints to encourage spatial diversity. Overall, our loss considerably improves the interpretability of the features, which now correspond to precise and anatomically meaningful landmarks. We demonstrate our three-fold loss in foetal rigid motion tracking and brain MRI affine registration tasks, where it not only outperforms state-of-the-art unsupervised strategies, but also bridges the gap with state-of-the-art supervised methods. Our code is available at https://github.com/BenBillot/spatial_regularisation.
CAMLD: Contrast-Agnostic Medical Landmark Detection with Consistency-Based Regularization
Nov 26, 2024Anatomical landmark detection in medical images is essential for various clinical and research applications, including disease diagnosis and surgical planning. However, manual landmark annotation is time-consuming and requires significant expertise. Existing deep learning (DL) methods often require large amounts of well-annotated data, which are costly to acquire. In this paper, we introduce CAMLD, a novel self-supervised DL framework for anatomical landmark detection in unlabeled scans with varying contrasts by using only a single reference example. To achieve this, we employed an inter-subject landmark consistency loss with an image registration loss while introducing a 3D convolution-based contrast augmentation strategy to promote model generalization to new contrasts. Additionally, we utilize an adaptive mixed loss function to schedule the contributions of different sub-tasks for optimal outcomes. We demonstrate the proposed method with the intricate task of MRI-based 3D brain landmark detection. With comprehensive experiments on four diverse clinical and public datasets, including both T1w and T2w MRI scans at different MRI field strengths, we demonstrate that CAMLD outperforms the state-of-the-art methods in terms of mean radial errors (MREs) and success detection rates (SDRs). Our framework provides a robust and accurate solution for anatomical landmark detection, reducing the need for extensively annotated datasets and generalizing well across different imaging contrasts. Our code will be publicly available at: https://github.com/HealthX-Lab/CAMLD.
A benchmark for 2D foetal brain ultrasound analysis
Jun 25, 2024Brain development involves a sequence of structural changes from early stages of the embryo until several months after birth. Currently, ultrasound is the established technique for screening due to its ability to acquire dynamic images in real-time without radiation and to its cost-efficiency. However, identifying abnormalities remains challenging due to the difficulty in interpreting foetal brain images. In this work we present a set of 104 2D foetal brain ultrasound images acquired during the 20th week of gestation that have been co-registered to a common space from a rough skull segmentation. The images are provided both on the original space and template space centred on the ellipses of all the subjects. Furthermore, the images have been annotated to highlight landmark points from structures of interest to analyse brain development. Both the final atlas template with probabilistic maps and the original images can be used to develop new segmentation techniques, test registration approaches for foetal brain ultrasound, extend our work to longitudinal datasets and to detect anomalies in new images.
Towards multi-modal anatomical landmark detection for ultrasound-guided brain tumor resection with contrastive learning
Jul 26, 2023



Homologous anatomical landmarks between medical scans are instrumental in quantitative assessment of image registration quality in various clinical applications, such as MRI-ultrasound registration for tissue shift correction in ultrasound-guided brain tumor resection. While manually identified landmark pairs between MRI and ultrasound (US) have greatly facilitated the validation of different registration algorithms for the task, the procedure requires significant expertise, labor, and time, and can be prone to inter- and intra-rater inconsistency. So far, many traditional and machine learning approaches have been presented for anatomical landmark detection, but they primarily focus on mono-modal applications. Unfortunately, despite the clinical needs, inter-modal/contrast landmark detection has very rarely been attempted. Therefore, we propose a novel contrastive learning framework to detect corresponding landmarks between MRI and intra-operative US scans in neurosurgery. Specifically, two convolutional neural networks were trained jointly to encode image features in MRI and US scans to help match the US image patch that contain the corresponding landmarks in the MRI. We developed and validated the technique using the public RESECT database. With a mean landmark detection accuracy of 5.88+-4.79 mm against 18.78+-4.77 mm with SIFT features, the proposed method offers promising results for MRI-US landmark detection in neurosurgical applications for the first time.
Automatic Landmark Detection and Registration of Brain Cortical Surfaces via Quasi-Conformal Geometry and Convolutional Neural Networks
Aug 15, 2022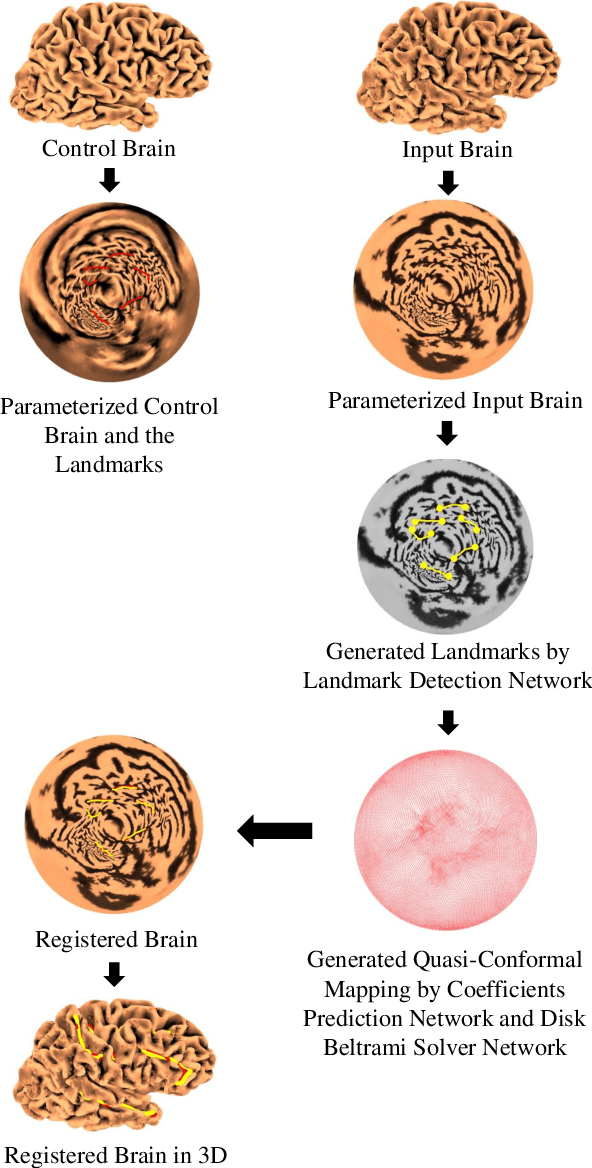
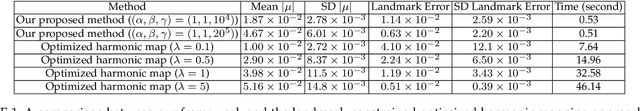
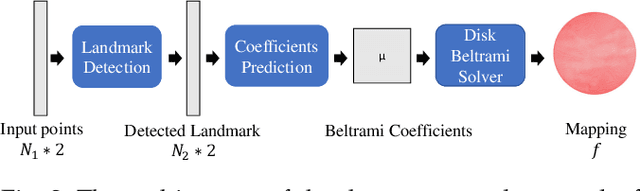

In medical imaging, surface registration is extensively used for performing systematic comparisons between anatomical structures, with a prime example being the highly convoluted brain cortical surfaces. To obtain a meaningful registration, a common approach is to identify prominent features on the surfaces and establish a low-distortion mapping between them with the feature correspondence encoded as landmark constraints. Prior registration works have primarily focused on using manually labeled landmarks and solving highly nonlinear optimization problems, which are time-consuming and hence hinder practical applications. In this work, we propose a novel framework for the automatic landmark detection and registration of brain cortical surfaces using quasi-conformal geometry and convolutional neural networks. We first develop a landmark detection network (LD-Net) that allows for the automatic extraction of landmark curves given two prescribed starting and ending points based on the surface geometry. We then utilize the detected landmarks and quasi-conformal theory for achieving the surface registration. Specifically, we develop a coefficient prediction network (CP-Net) for predicting the Beltrami coefficients associated with the desired landmark-based registration and a mapping network called the disk Beltrami solver network (DBS-Net) for generating quasi-conformal mappings from the predicted Beltrami coefficients, with the bijectivity guaranteed by quasi-conformal theory. Experimental results are presented to demonstrate the effectiveness of our proposed framework. Altogether, our work paves a new way for surface-based morphometry and medical shape analysis.
Towards Practical Application of Deep Learning in Diagnosis of Alzheimer's Disease
Dec 08, 2022Accurate diagnosis of Alzheimer's disease (AD) is both challenging and time consuming. With a systematic approach for early detection and diagnosis of AD, steps can be taken towards the treatment and prevention of the disease. This study explores the practical application of deep learning models for diagnosis of AD. Due to computational complexity, large training times and limited availability of labelled dataset, a 3D full brain CNN (convolutional neural network) is not commonly used, and researchers often prefer 2D CNN variants. In this study, full brain 3D version of well-known 2D CNNs were designed, trained and tested for diagnosis of various stages of AD. Deep learning approach shows good performance in differentiating various stages of AD for more than 1500 full brain volumes. Along with classification, the deep learning model is capable of extracting features which are key in differentiating the various categories. The extracted features align with meaningful anatomical landmarks, that are currently considered important in identification of AD by experts. An ensemble of all the algorithm was also tested and the performance of the ensemble algorithm was superior to any individual algorithm, further improving diagnosis ability. The 3D versions of the trained CNNs and their ensemble have the potential to be incorporated in software packages that can be used by physicians/radiologists to assist them in better diagnosis of AD.
Simulating Realistic MRI variations to Improve Deep Learning model and visual explanations using GradCAM
Nov 01, 2021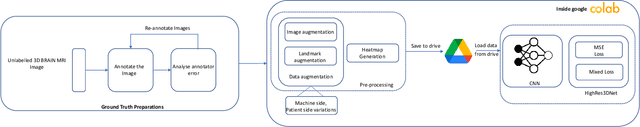
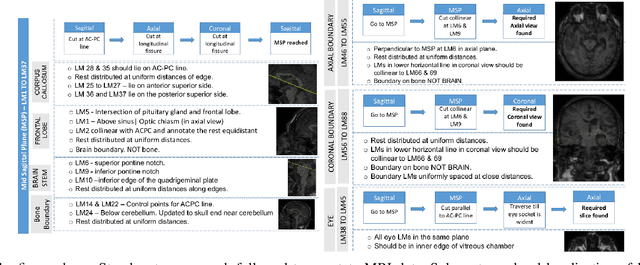
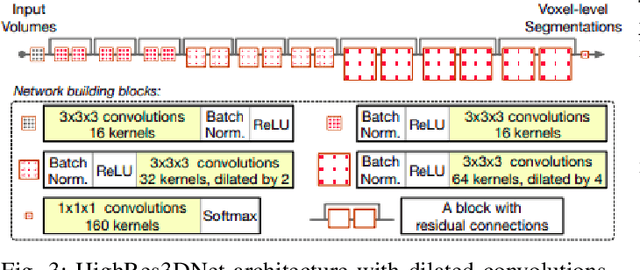
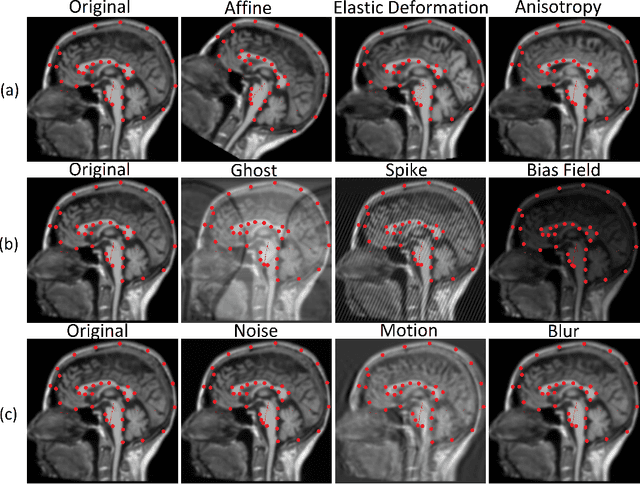
In the medical field, landmark detection in MRI plays an important role in reducing medical technician efforts in tasks like scan planning, image registration, etc. First, 88 landmarks spread across the brain anatomy in the three respective views -- sagittal, coronal, and axial are manually annotated, later guidelines from the expert clinical technicians are taken sub-anatomy-wise, for better localization of the existing landmarks, in order to identify and locate the important atlas landmarks even in oblique scans. To overcome limited data availability, we implement realistic data augmentation to generate synthetic 3D volumetric data. We use a modified HighRes3DNet model for solving brain MRI volumetric landmark detection problem. In order to visually explain our trained model on unseen data, and discern a stronger model from a weaker model, we implement Gradient-weighted Class Activation Mapping (Grad-CAM) which produces a coarse localization map highlighting the regions the model is focusing. Our experiments show that the proposed method shows favorable results, and the overall pipeline can be extended to a variable number of landmarks and other anatomies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge