Zizhi Chen
Fusing Pixels and Genes: Spatially-Aware Learning in Computational Pathology
Feb 15, 2026Abstract:Recent years have witnessed remarkable progress in multimodal learning within computational pathology. Existing models primarily rely on vision and language modalities; however, language alone lacks molecular specificity and offers limited pathological supervision, leading to representational bottlenecks. In this paper, we propose STAMP, a Spatial Transcriptomics-Augmented Multimodal Pathology representation learning framework that integrates spatially-resolved gene expression profiles to enable molecule-guided joint embedding of pathology images and transcriptomic data. Our study shows that self-supervised, gene-guided training provides a robust and task-agnostic signal for learning pathology image representations. Incorporating spatial context and multi-scale information further enhances model performance and generalizability. To support this, we constructed SpaVis-6M, the largest Visium-based spatial transcriptomics dataset to date, and trained a spatially-aware gene encoder on this resource. Leveraging hierarchical multi-scale contrastive alignment and cross-scale patch localization mechanisms, STAMP effectively aligns spatial transcriptomics with pathology images, capturing spatial structure and molecular variation. We validate STAMP across six datasets and four downstream tasks, where it consistently achieves strong performance. These results highlight the value and necessity of integrating spatially resolved molecular supervision for advancing multimodal learning in computational pathology. The code is included in the supplementary materials. The pretrained weights and SpaVis-6M are available at: https://github.com/Hanminghao/STAMP.
Beyond Pixel Simulation: Pathology Image Generation via Diagnostic Semantic Tokens and Prototype Control
Dec 24, 2025



Abstract:In computational pathology, understanding and generation have evolved along disparate paths: advanced understanding models already exhibit diagnostic-level competence, whereas generative models largely simulate pixels. Progress remains hindered by three coupled factors: the scarcity of large, high-quality image-text corpora; the lack of precise, fine-grained semantic control, which forces reliance on non-semantic cues; and terminological heterogeneity, where diverse phrasings for the same diagnostic concept impede reliable text conditioning. We introduce UniPath, a semantics-driven pathology image generation framework that leverages mature diagnostic understanding to enable controllable generation. UniPath implements Multi-Stream Control: a Raw-Text stream; a High-Level Semantics stream that uses learnable queries to a frozen pathology MLLM to distill paraphrase-robust Diagnostic Semantic Tokens and to expand prompts into diagnosis-aware attribute bundles; and a Prototype stream that affords component-level morphological control via a prototype bank. On the data front, we curate a 2.65M image-text corpus and a finely annotated, high-quality 68K subset to alleviate data scarcity. For a comprehensive assessment, we establish a four-tier evaluation hierarchy tailored to pathology. Extensive experiments demonstrate UniPath's SOTA performance, including a Patho-FID of 80.9 (51% better than the second-best) and fine-grained semantic control achieving 98.7% of the real-image. The meticulously curated datasets, complete source code, and pre-trained model weights developed in this study will be made openly accessible to the public.
Forging a Dynamic Memory: Retrieval-Guided Continual Learning for Generalist Medical Foundation Models
Dec 15, 2025Abstract:Multimodal biomedical Vision-Language Models (VLMs) exhibit immense potential in the field of Continual Learning (CL). However, they confront a core dilemma: how to preserve fine-grained intra-modality features while bridging the significant domain gap across different modalities. To address this challenge, we propose a comprehensive framework. Leveraging our 18-million multimodal and comprehensive medical retrieval database derived from PubMed scientific papers, we pioneer the integration of Retrieval-Augmented Generation (RAG) into CL. Specifically, we employ a multi-modal, multi-layer RAG system that provides real-time guidance for model fine-tuning through dynamic, on-demand knowledge retrieval. Building upon this, we introduce a dynamic knowledge distillation framework. This framework precisely resolves the aforementioned core dilemma by dynamically modulating the importance of the parameter space, the granularity of the distilled knowledge, and the data distribution of the reference dataset in accordance with the required level of detail. To thoroughly validate the clinical value of our strategy, we have designed a more rigorous \textbf{M}edical Generalist Task Incremental Learning (MGTIL) benchmark. This benchmark is engineered to simultaneously evaluate the model's capacity for adaptation to significant domain shifts, retention of subtle intra-domain features, and real-time learning of novel and complex medical tasks. Extensive experimental results demonstrate that our proposed method achieves state-of-the-art (SOTA) performance across all metrics. The code is provided in the supplementary materials.
FysicsWorld: A Unified Full-Modality Benchmark for Any-to-Any Understanding, Generation, and Reasoning
Dec 14, 2025Abstract:Despite rapid progress in multimodal large language models (MLLMs) and emerging omni-modal architectures, current benchmarks remain limited in scope and integration, suffering from incomplete modality coverage, restricted interaction to text-centric outputs, and weak interdependence and complementarity among modalities. To bridge these gaps, we introduce FysicsWorld, the first unified full-modality benchmark that supports bidirectional input-output across image, video, audio, and text, enabling comprehensive any-to-any evaluation across understanding, generation, and reasoning. FysicsWorld encompasses 16 primary tasks and 3,268 curated samples, aggregated from over 40 high-quality sources and covering a rich set of open-domain categories with diverse question types. We also propose the Cross-Modal Complementarity Screening (CMCS) strategy integrated in a systematic data construction framework that produces omni-modal data for spoken interaction and fusion-dependent cross-modal reasoning. Through a comprehensive evaluation of over 30 state-of-the-art baselines, spanning MLLMs, modality-specific models, unified understanding-generation models, and omni-modal language models, FysicsWorld exposes the performance disparities and limitations across models in understanding, generation, and reasoning. Our benchmark establishes a unified foundation and strong baselines for evaluating and advancing next-generation full-modality architectures.
PersonaAnimator: Personalized Motion Transfer from Unconstrained Videos
Aug 27, 2025Abstract:Recent advances in motion generation show remarkable progress. However, several limitations remain: (1) Existing pose-guided character motion transfer methods merely replicate motion without learning its style characteristics, resulting in inexpressive characters. (2) Motion style transfer methods rely heavily on motion capture data, which is difficult to obtain. (3) Generated motions sometimes violate physical laws. To address these challenges, this paper pioneers a new task: Video-to-Video Motion Personalization. We propose a novel framework, PersonaAnimator, which learns personalized motion patterns directly from unconstrained videos. This enables personalized motion transfer. To support this task, we introduce PersonaVid, the first video-based personalized motion dataset. It contains 20 motion content categories and 120 motion style categories. We further propose a Physics-aware Motion Style Regularization mechanism to enforce physical plausibility in the generated motions. Extensive experiments show that PersonaAnimator outperforms state-of-the-art motion transfer methods and sets a new benchmark for the Video-to-Video Motion Personalization task.
VLM-based Prompts as the Optimal Assistant for Unpaired Histopathology Virtual Staining
Apr 22, 2025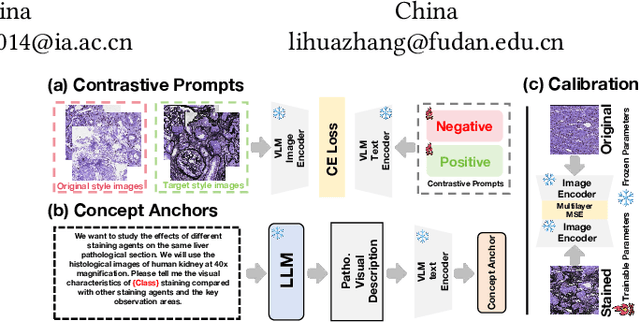



Abstract:In histopathology, tissue sections are typically stained using common H&E staining or special stains (MAS, PAS, PASM, etc.) to clearly visualize specific tissue structures. The rapid advancement of deep learning offers an effective solution for generating virtually stained images, significantly reducing the time and labor costs associated with traditional histochemical staining. However, a new challenge arises in separating the fundamental visual characteristics of tissue sections from the visual differences induced by staining agents. Additionally, virtual staining often overlooks essential pathological knowledge and the physical properties of staining, resulting in only style-level transfer. To address these issues, we introduce, for the first time in virtual staining tasks, a pathological vision-language large model (VLM) as an auxiliary tool. We integrate contrastive learnable prompts, foundational concept anchors for tissue sections, and staining-specific concept anchors to leverage the extensive knowledge of the pathological VLM. This approach is designed to describe, frame, and enhance the direction of virtual staining. Furthermore, we have developed a data augmentation method based on the constraints of the VLM. This method utilizes the VLM's powerful image interpretation capabilities to further integrate image style and structural information, proving beneficial in high-precision pathological diagnostics. Extensive evaluations on publicly available multi-domain unpaired staining datasets demonstrate that our method can generate highly realistic images and enhance the accuracy of downstream tasks, such as glomerular detection and segmentation. Our code is available at: https://github.com/CZZZZZZZZZZZZZZZZZ/VPGAN-HARBOR
VGAT: A Cancer Survival Analysis Framework Transitioning from Generative Visual Question Answering to Genomic Reconstruction
Mar 25, 2025Abstract:Multimodal learning combining pathology images and genomic sequences enhances cancer survival analysis but faces clinical implementation barriers due to limited access to genomic sequencing in under-resourced regions. To enable survival prediction using only whole-slide images (WSI), we propose the Visual-Genomic Answering-Guided Transformer (VGAT), a framework integrating Visual Question Answering (VQA) techniques for genomic modality reconstruction. By adapting VQA's text feature extraction approach, we derive stable genomic representations that circumvent dimensionality challenges in raw genomic data. Simultaneously, a cluster-based visual prompt module selectively enhances discriminative WSI patches, addressing noise from unfiltered image regions. Evaluated across five TCGA datasets, VGAT outperforms existing WSI-only methods, demonstrating the viability of genomic-informed inference without sequencing. This approach bridges multimodal research and clinical feasibility in resource-constrained settings. The code link is https://github.com/CZZZZZZZZZZZZZZZZZ/VGAT.
Towards Unified Molecule-Enhanced Pathology Image Representation Learning via Integrating Spatial Transcriptomics
Dec 01, 2024



Abstract:Recent advancements in multimodal pre-training models have significantly advanced computational pathology. However, current approaches predominantly rely on visual-language models, which may impose limitations from a molecular perspective and lead to performance bottlenecks. Here, we introduce a Unified Molecule-enhanced Pathology Image REpresentationn Learning framework (UMPIRE). UMPIRE aims to leverage complementary information from gene expression profiles to guide the multimodal pre-training, enhancing the molecular awareness of pathology image representation learning. We demonstrate that this molecular perspective provides a robust, task-agnostic training signal for learning pathology image embeddings. Due to the scarcity of paired data, approximately 4 million entries of spatial transcriptomics gene expression were collected to train the gene encoder. By leveraging powerful pre-trained encoders, UMPIRE aligns the encoders across over 697K pathology image-gene expression pairs. The performance of UMPIRE is demonstrated across various molecular-related downstream tasks, including gene expression prediction, spot classification, and mutation state prediction in whole slide images. Our findings highlight the effectiveness of multimodal data integration and open new avenues for exploring computational pathology enhanced by molecular perspectives. The code and pre-trained weights are available at https://github.com/Hanminghao/UMPIRE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge