Yuyue Zhou
FlexICL: A Flexible Visual In-context Learning Framework for Elbow and Wrist Ultrasound Segmentation
Oct 30, 2025Abstract:Elbow and wrist fractures are the most common fractures in pediatric populations. Automatic segmentation of musculoskeletal structures in ultrasound (US) can improve diagnostic accuracy and treatment planning. Fractures appear as cortical defects but require expert interpretation. Deep learning (DL) can provide real-time feedback and highlight key structures, helping lightly trained users perform exams more confidently. However, pixel-wise expert annotations for training remain time-consuming and costly. To address this challenge, we propose FlexICL, a novel and flexible in-context learning (ICL) framework for segmenting bony regions in US images. We apply it to an intra-video segmentation setting, where experts annotate only a small subset of frames, and the model segments unseen frames. We systematically investigate various image concatenation techniques and training strategies for visual ICL and introduce novel concatenation methods that significantly enhance model performance with limited labeled data. By integrating multiple augmentation strategies, FlexICL achieves robust segmentation performance across four wrist and elbow US datasets while requiring only 5% of the training images. It outperforms state-of-the-art visual ICL models like Painter, MAE-VQGAN, and conventional segmentation models like U-Net and TransUNet by 1-27% Dice coefficient on 1,252 US sweeps. These initial results highlight the potential of FlexICL as an efficient and scalable solution for US image segmentation well suited for medical imaging use cases where labeled data is scarce.
Phased Array Calibration based on Rotating-Element Harmonic Electric-Field Vector with Time Modulation
Apr 17, 2025



Abstract:Calibration is crucial for ensuring the performance of phased array since amplitude-phase imbalance between elements results in significant performance degradation. While amplitude-only calibration methods offer advantages when phase measurements are impractical, conventional approaches face two key challenges: they typically require high-resolution phase shifters and remain susceptible to phase errors inherent in these components. To overcome these limitations, we propose a Rotating element Harmonic Electric-field Vector (RHEV) strategy, which enables precise calibration through time modulation principles. The proposed technique functions as follows. Two 1-bit phase shifters are periodically phase-switched at the same frequency, each generating corresponding harmonics. By adjusting the relative delay between their modulation timings, the phase difference between the $+1$st harmonics produced by the two elements can be precisely controlled, utilizing the time-shift property of the Fourier transform. Furthermore, the +1st harmonic generated by sequential modulation of individual elements exhibits a linear relationship with the amplitude of the modulated element, enabling amplitude ambiguity resolution. The proposed RHEV-based calibration method generates phase shifts through relative timing delays rather than physical phase shifter adjustments, rendering it less susceptible to phase shift errors. Additionally, since the calibration process exclusively utilizes the $+1$st harmonic, which is produced solely by the modulated unit, the method demonstrates consistent performance regardless of array size. Extensive numerical simulations, practical in-channel and over-the-air (OTA) calibration experiments demonstrate the effectiveness and distinct advantages of the proposed method.
Sam2Rad: A Segmentation Model for Medical Images with Learnable Prompts
Sep 10, 2024



Abstract:Foundation models like the segment anything model require high-quality manual prompts for medical image segmentation, which is time-consuming and requires expertise. SAM and its variants often fail to segment structures in ultrasound (US) images due to domain shift. We propose Sam2Rad, a prompt learning approach to adapt SAM and its variants for US bone segmentation without human prompts. It introduces a prompt predictor network (PPN) with a cross-attention module to predict prompt embeddings from image encoder features. PPN outputs bounding box and mask prompts, and 256-dimensional embeddings for regions of interest. The framework allows optional manual prompting and can be trained end-to-end using parameter-efficient fine-tuning (PEFT). Sam2Rad was tested on 3 musculoskeletal US datasets: wrist (3822 images), rotator cuff (1605 images), and hip (4849 images). It improved performance across all datasets without manual prompts, increasing Dice scores by 2-7% for hip/wrist and up to 33% for shoulder data. Sam2Rad can be trained with as few as 10 labeled images and is compatible with any SAM architecture for automatic segmentation.
A Simple Framework Uniting Visual In-context Learning with Masked Image Modeling to Improve Ultrasound Segmentation
Mar 08, 2024



Abstract:Conventional deep learning models deal with images one-by-one, requiring costly and time-consuming expert labeling in the field of medical imaging, and domain-specific restriction limits model generalizability. Visual in-context learning (ICL) is a new and exciting area of research in computer vision. Unlike conventional deep learning, ICL emphasizes the model's ability to adapt to new tasks based on given examples quickly. Inspired by MAE-VQGAN, we proposed a new simple visual ICL method called SimICL, combining visual ICL pairing images with masked image modeling (MIM) designed for self-supervised learning. We validated our method on bony structures segmentation in a wrist ultrasound (US) dataset with limited annotations, where the clinical objective was to segment bony structures to help with further fracture detection. We used a test set containing 3822 images from 18 patients for bony region segmentation. SimICL achieved an remarkably high Dice coeffient (DC) of 0.96 and Jaccard Index (IoU) of 0.92, surpassing state-of-the-art segmentation and visual ICL models (a maximum DC 0.86 and IoU 0.76), with SimICL DC and IoU increasing up to 0.10 and 0.16. This remarkably high agreement with limited manual annotations indicates SimICL could be used for training AI models even on small US datasets. This could dramatically decrease the human expert time required for image labeling compared to conventional approaches, and enhance the real-world use of AI assistance in US image analysis.
Application Of Vision-Language Models For Assessing Osteoarthritis Disease Severity
Jan 12, 2024Abstract:Osteoarthritis (OA) poses a global health challenge, demanding precise diagnostic methods. Current radiographic assessments are time consuming and prone to variability, prompting the need for automated solutions. The existing deep learning models for OA assessment are unimodal single task systems and they don't incorporate relevant text information such as patient demographics, disease history, or physician reports. This study investigates employing Vision Language Processing (VLP) models to predict OA severity using Xray images and corresponding reports. Our method leverages Xray images of the knee and diverse report templates generated from tabular OA scoring values to train a CLIP (Contrastive Language Image PreTraining) style VLP model. Furthermore, we incorporate additional contrasting captions to enforce the model to discriminate between positive and negative reports. Results demonstrate the efficacy of these models in learning text image representations and their contextual relationships, showcase potential advancement in OA assessment, and establish a foundation for specialized vision language models in medical contexts.
Self-supervised TransUNet for Ultrasound regional segmentation of the distal radius in children
Sep 18, 2023



Abstract:Supervised deep learning offers great promise to automate analysis of medical images from segmentation to diagnosis. However, their performance highly relies on the quality and quantity of the data annotation. Meanwhile, curating large annotated datasets for medical images requires a high level of expertise, which is time-consuming and expensive. Recently, to quench the thirst for large data sets with high-quality annotation, self-supervised learning (SSL) methods using unlabeled domain-specific data, have attracted attention. Therefore, designing an SSL method that relies on minimal quantities of labeled data has far-reaching significance in medical images. This paper investigates the feasibility of deploying the Masked Autoencoder for SSL (SSL-MAE) of TransUNet, for segmenting bony regions from children's wrist ultrasound scans. We found that changing the embedding and loss function in SSL-MAE can produce better downstream results compared to the original SSL-MAE. In addition, we determined that only pretraining TransUNet embedding and encoder with SSL-MAE does not work as well as TransUNet without SSL-MAE pretraining on downstream segmentation tasks.
Improving Tuberculosis (TB) Prediction using Synthetically Generated Computed Tomography (CT) Images
Sep 23, 2021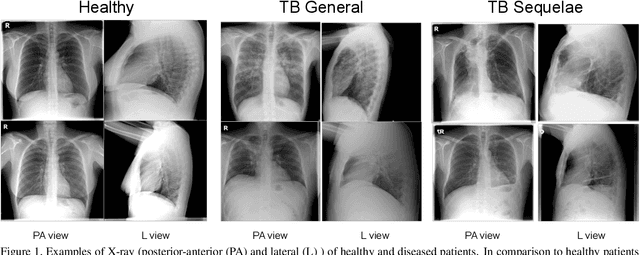
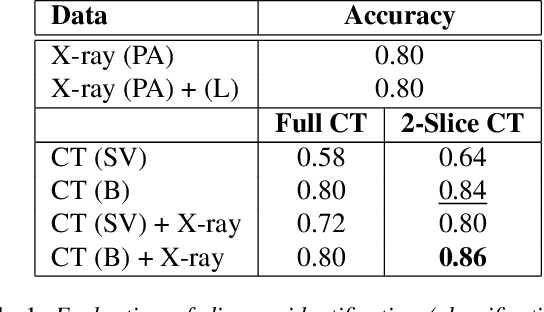
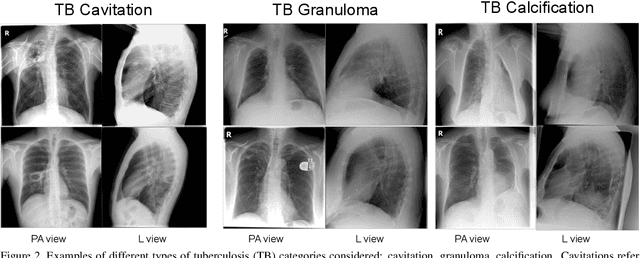
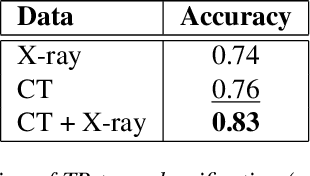
Abstract:The evaluation of infectious disease processes on radiologic images is an important and challenging task in medical image analysis. Pulmonary infections can often be best imaged and evaluated through computed tomography (CT) scans, which are often not available in low-resource environments and difficult to obtain for critically ill patients. On the other hand, X-ray, a different type of imaging procedure, is inexpensive, often available at the bedside and more widely available, but offers a simpler, two dimensional image. We show that by relying on a model that learns to generate CT images from X-rays synthetically, we can improve the automatic disease classification accuracy and provide clinicians with a different look at the pulmonary disease process. Specifically, we investigate Tuberculosis (TB), a deadly bacterial infectious disease that predominantly affects the lungs, but also other organ systems. We show that relying on synthetically generated CT improves TB identification by 7.50% and distinguishes TB properties up to 12.16% better than the X-ray baseline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge