Abhilash Hareendranathan
Retuve: Automated Multi-Modality Analysis of Hip Dysplasia with Open Source AI
Apr 08, 2025Abstract:Developmental dysplasia of the hip (DDH) poses significant diagnostic challenges, hindering timely intervention. Current screening methodologies lack standardization, and AI-driven studies suffer from reproducibility issues due to limited data and code availability. To address these limitations, we introduce Retuve, an open-source framework for multi-modality DDH analysis, encompassing both ultrasound (US) and X-ray imaging. Retuve provides a complete and reproducible workflow, offering open datasets comprising expert-annotated US and X-ray images, pre-trained models with training code and weights, and a user-friendly Python Application Programming Interface (API). The framework integrates segmentation and landmark detection models, enabling automated measurement of key diagnostic parameters such as the alpha angle and acetabular index. By adhering to open-source principles, Retuve promotes transparency, collaboration, and accessibility in DDH research. This initiative has the potential to democratize DDH screening, facilitate early diagnosis, and ultimately improve patient outcomes by enabling widespread screening and early intervention. The GitHub repository/code can be found here: https://github.com/radoss-org/retuve
Reinforcement Learning for Ultrasound Image Analysis A Comprehensive Review of Advances and Applications
Feb 20, 2025Abstract:Over the last decade, the use of machine learning (ML) approaches in medicinal applications has increased manifold. Most of these approaches are based on deep learning, which aims to learn representations from grid data (like medical images). However, reinforcement learning (RL) applications in medicine are relatively less explored. Medical applications often involve a sequence of subtasks that form a diagnostic pipeline, and RL is uniquely suited to optimize over such sequential decision-making tasks. Ultrasound (US) image analysis is a quintessential example of such a sequential decision-making task, where the raw signal captured by the US transducer undergoes a series of signal processing and image post-processing steps, generally leading to a diagnostic suggestion. The application of RL in US remains limited. Deep Reinforcement Learning (DRL), that combines deep learning and RL, holds great promise in optimizing these pipelines by enabling intelligent and sequential decision-making. This review paper surveys the applications of RL in US over the last decade. We provide a succinct overview of the theoretic framework of RL and its application in US image processing and review existing work in each aspect of the image analysis pipeline. A comprehensive search of Scopus filtered on relevance yielded 14 papers most relevant to this topic. These papers were further categorized based on their target applications image classification, image segmentation, image enhancement, video summarization, and auto navigation and path planning. We also examined the type of RL approach used in each publication. Finally, we discuss key areas in healthcare where DRL approaches in US could be used for sequential decision-making. We analyze the opportunities, challenges, and limitations, providing insights into the future potential of DRL in US image analysis.
Sam2Rad: A Segmentation Model for Medical Images with Learnable Prompts
Sep 10, 2024



Abstract:Foundation models like the segment anything model require high-quality manual prompts for medical image segmentation, which is time-consuming and requires expertise. SAM and its variants often fail to segment structures in ultrasound (US) images due to domain shift. We propose Sam2Rad, a prompt learning approach to adapt SAM and its variants for US bone segmentation without human prompts. It introduces a prompt predictor network (PPN) with a cross-attention module to predict prompt embeddings from image encoder features. PPN outputs bounding box and mask prompts, and 256-dimensional embeddings for regions of interest. The framework allows optional manual prompting and can be trained end-to-end using parameter-efficient fine-tuning (PEFT). Sam2Rad was tested on 3 musculoskeletal US datasets: wrist (3822 images), rotator cuff (1605 images), and hip (4849 images). It improved performance across all datasets without manual prompts, increasing Dice scores by 2-7% for hip/wrist and up to 33% for shoulder data. Sam2Rad can be trained with as few as 10 labeled images and is compatible with any SAM architecture for automatic segmentation.
Application Of Vision-Language Models For Assessing Osteoarthritis Disease Severity
Jan 12, 2024Abstract:Osteoarthritis (OA) poses a global health challenge, demanding precise diagnostic methods. Current radiographic assessments are time consuming and prone to variability, prompting the need for automated solutions. The existing deep learning models for OA assessment are unimodal single task systems and they don't incorporate relevant text information such as patient demographics, disease history, or physician reports. This study investigates employing Vision Language Processing (VLP) models to predict OA severity using Xray images and corresponding reports. Our method leverages Xray images of the knee and diverse report templates generated from tabular OA scoring values to train a CLIP (Contrastive Language Image PreTraining) style VLP model. Furthermore, we incorporate additional contrasting captions to enforce the model to discriminate between positive and negative reports. Results demonstrate the efficacy of these models in learning text image representations and their contextual relationships, showcase potential advancement in OA assessment, and establish a foundation for specialized vision language models in medical contexts.
Weakly Supervised Medical Image Segmentation With Soft Labels and Noise Robust Loss
Sep 16, 2022
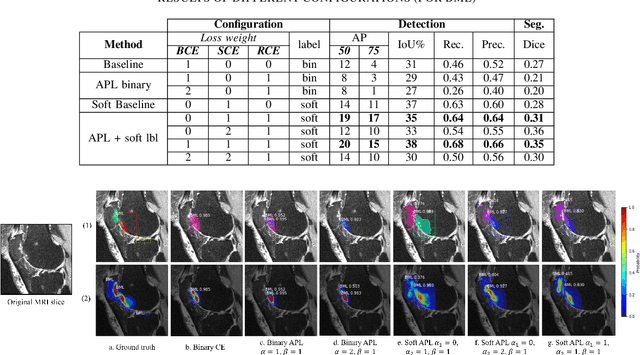
Abstract:Recent advances in deep learning algorithms have led to significant benefits for solving many medical image analysis problems. Training deep learning models commonly requires large datasets with expert-labeled annotations. However, acquiring expert-labeled annotation is not only expensive but also is subjective, error-prone, and inter-/intra- observer variability introduces noise to labels. This is particularly a problem when using deep learning models for segmenting medical images due to the ambiguous anatomical boundaries. Image-based medical diagnosis tools using deep learning models trained with incorrect segmentation labels can lead to false diagnoses and treatment suggestions. Multi-rater annotations might be better suited to train deep learning models with small training sets compared to single-rater annotations. The aim of this paper was to develop and evaluate a method to generate probabilistic labels based on multi-rater annotations and anatomical knowledge of the lesion features in MRI and a method to train segmentation models using probabilistic labels using normalized active-passive loss as a "noise-tolerant loss" function. The model was evaluated by comparing it to binary ground truth for 17 knees MRI scans for clinical segmentation and detection of bone marrow lesions (BML). The proposed method successfully improved precision 14, recall 22, and Dice score 8 percent compared to a binary cross-entropy loss function. Overall, the results of this work suggest that the proposed normalized active-passive loss using soft labels successfully mitigated the effects of noisy labels.
Self-Supervised-RCNN for Medical Image Segmentation with Limited Data Annotation
Jul 17, 2022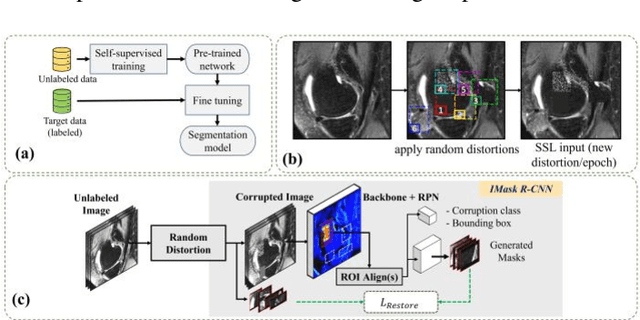
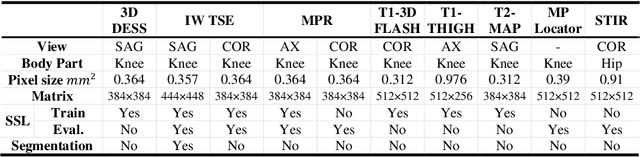
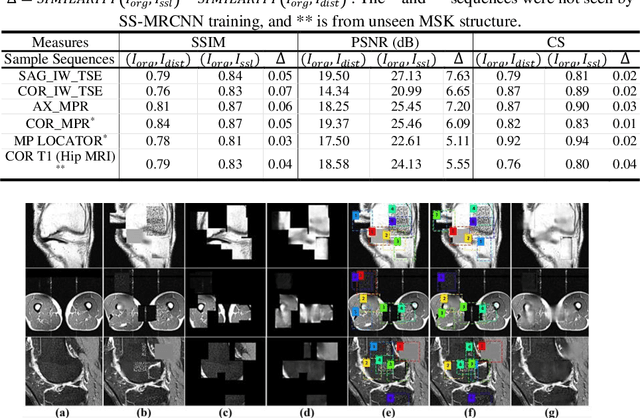
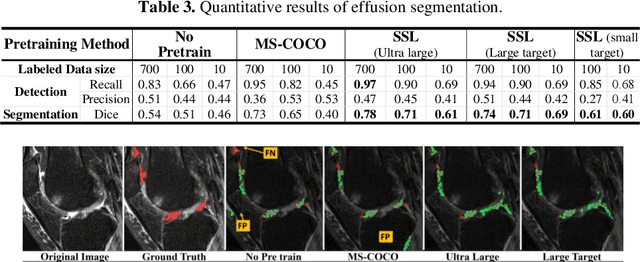
Abstract:Many successful methods developed for medical image analysis that are based on machine learning use supervised learning approaches, which often require large datasets annotated by experts to achieve high accuracy. However, medical data annotation is time-consuming and expensive, especially for segmentation tasks. To solve the problem of learning with limited labeled medical image data, an alternative deep learning training strategy based on self-supervised pretraining on unlabeled MRI scans is proposed in this work. Our pretraining approach first, randomly applies different distortions to random areas of unlabeled images and then predicts the type of distortions and loss of information. To this aim, an improved version of Mask-RCNN architecture has been adapted to localize the distortion location and recover the original image pixels. The effectiveness of the proposed method for segmentation tasks in different pre-training and fine-tuning scenarios is evaluated based on the Osteoarthritis Initiative dataset. Using this self-supervised pretraining method improved the Dice score by 20% compared to training from scratch. The proposed self-supervised learning is simple, effective, and suitable for different ranges of medical image analysis tasks including anomaly detection, segmentation, and classification.
Improved-Mask R-CNN: Towards an Accurate Generic MSK MRI instance segmentation platform
Jul 27, 2021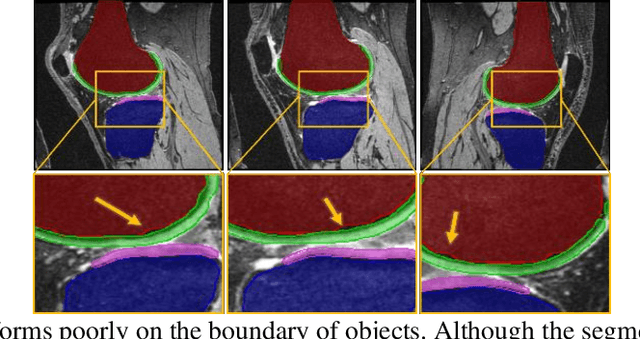
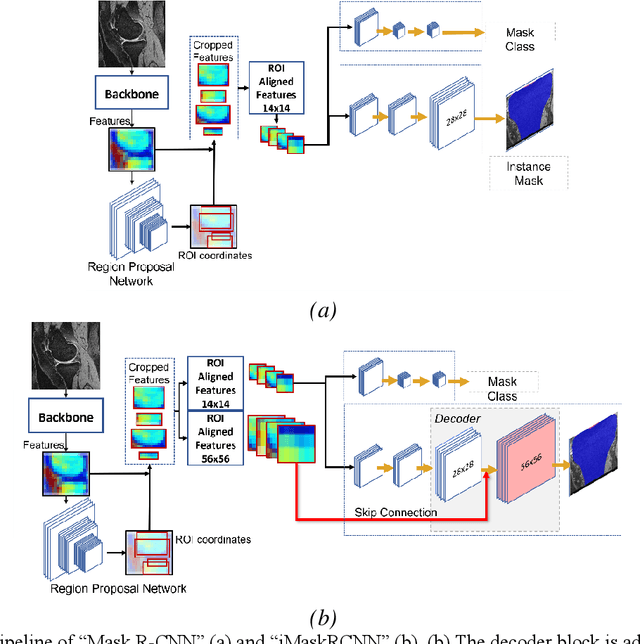
Abstract:Objective assessment of Magnetic Resonance Imaging (MRI) scans of osteoarthritis (OA) can address the limitation of the current OA assessment. Segmentation of bone, cartilage, and joint fluid is necessary for the OA objective assessment. Most of the proposed segmentation methods are not performing instance segmentation and suffer from class imbalance problems. This study deployed Mask R-CNN instance segmentation and improved it (improved-Mask R-CNN (iMaskRCNN)) to obtain a more accurate generalized segmentation for OA-associated tissues. Training and validation of the method were performed using 500 MRI knees from the Osteoarthritis Initiative (OAI) dataset and 97 MRI scans of patients with symptomatic hip OA. Three modifications to Mask R-CNN yielded the iMaskRCNN: adding a 2nd ROIAligned block, adding an extra decoder layer to the mask-header, and connecting them by a skip connection. The results were assessed using Hausdorff distance, dice score, and coefficients of variation (CoV). The iMaskRCNN led to improved bone and cartilage segmentation compared to Mask RCNN as indicated with the increase in dice score from 95% to 98% for the femur, 95% to 97% for tibia, 71% to 80% for femoral cartilage, and 81% to 82% for tibial cartilage. For the effusion detection, dice improved with iMaskRCNN 72% versus MaskRCNN 71%. The CoV values for effusion detection between Reader1 and Mask R-CNN (0.33), Reader1 and iMaskRCNN (0.34), Reader2 and Mask R-CNN (0.22), Reader2 and iMaskRCNN (0.29) are close to CoV between two readers (0.21), indicating a high agreement between the human readers and both Mask R-CNN and iMaskRCNN. Mask R-CNN and iMaskRCNN can reliably and simultaneously extract different scale articular tissues involved in OA, forming the foundation for automated assessment of OA. The iMaskRCNN results show that the modification improved the network performance around the edges.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge