Xuhui Huang
Department of Chemistry, Center of System Biology and Human Health, State Key Laboratory of Molecular Neuroscience, Hong Kong University of Science and Technology
AutoSDT: Scaling Data-Driven Discovery Tasks Toward Open Co-Scientists
Jun 09, 2025



Abstract:Despite long-standing efforts in accelerating scientific discovery with AI, building AI co-scientists remains challenging due to limited high-quality data for training and evaluation. To tackle this data scarcity issue, we present AutoSDT, an automatic pipeline that collects high-quality coding tasks in real-world data-driven discovery workflows. AutoSDT leverages the coding capabilities and parametric knowledge of LLMs to search for diverse sources, select ecologically valid tasks, and synthesize accurate task instructions and code solutions. Using our pipeline, we construct AutoSDT-5K, a dataset of 5,404 coding tasks for data-driven discovery that covers four scientific disciplines and 756 unique Python packages. To the best of our knowledge, AutoSDT-5K is the only automatically collected and the largest open dataset for data-driven scientific discovery. Expert feedback on a subset of 256 tasks shows the effectiveness of AutoSDT: 93% of the collected tasks are ecologically valid, and 92.2% of the synthesized programs are functionally correct. Trained on AutoSDT-5K, the Qwen2.5-Coder-Instruct LLM series, dubbed AutoSDT-Coder, show substantial improvement on two challenging data-driven discovery benchmarks, ScienceAgentBench and DiscoveryBench. Most notably, AutoSDT-Coder-32B reaches the same level of performance as GPT-4o on ScienceAgentBench with a success rate of 7.8%, doubling the performance of its base model. On DiscoveryBench, it lifts the hypothesis matching score to 8.1, bringing a 17.4% relative improvement and closing the gap between open-weight models and GPT-4o.
Improving Transformer Based Line Segment Detection with Matched Predicting and Re-ranking
Feb 25, 2025Abstract:Classical Transformer-based line segment detection methods have delivered impressive results. However, we observe that some accurately detected line segments are assigned low confidence scores during prediction, causing them to be ranked lower and potentially suppressed. Additionally, these models often require prolonged training periods to achieve strong performance, largely due to the necessity of bipartite matching. In this paper, we introduce RANK-LETR, a novel Transformer-based line segment detection method. Our approach leverages learnable geometric information to refine the ranking of predicted line segments by enhancing the confidence scores of high-quality predictions in a posterior verification step. We also propose a new line segment proposal method, wherein the feature point nearest to the centroid of the line segment directly predicts the location, significantly improving training efficiency and stability. Moreover, we introduce a line segment ranking loss to stabilize rankings during training, thereby enhancing the generalization capability of the model. Experimental results demonstrate that our method outperforms other Transformer-based and CNN-based approaches in prediction accuracy while requiring fewer training epochs than previous Transformer-based models.
ScienceAgentBench: Toward Rigorous Assessment of Language Agents for Data-Driven Scientific Discovery
Oct 07, 2024



Abstract:The advancements of language language models (LLMs) have piqued growing interest in developing LLM-based language agents to automate scientific discovery end-to-end, which has sparked both excitement and skepticism about the true capabilities of such agents. In this work, we argue that for an agent to fully automate scientific discovery, it must be able to complete all essential tasks in the workflow. Thus, we call for rigorous assessment of agents on individual tasks in a scientific workflow before making bold claims on end-to-end automation. To this end, we present ScienceAgentBench, a new benchmark for evaluating language agents for data-driven scientific discovery. To ensure the scientific authenticity and real-world relevance of our benchmark, we extract 102 tasks from 44 peer-reviewed publications in four disciplines and engage nine subject matter experts to validate them. We unify the target output for every task to a self-contained Python program file and employ an array of evaluation metrics to examine the generated programs, execution results, and costs. Each task goes through multiple rounds of manual validation by annotators and subject matter experts to ensure its annotation quality and scientific plausibility. We also propose two effective strategies to mitigate data contamination concerns. Using our benchmark, we evaluate five open-weight and proprietary LLMs, each with three frameworks: direct prompting, OpenHands, and self-debug. Given three attempts for each task, the best-performing agent can only solve 32.4% of the tasks independently and 34.3% with expert-provided knowledge. These results underscore the limited capacities of current language agents in generating code for data-driven discovery, let alone end-to-end automation for scientific research.
Ternary Spike: Learning Ternary Spikes for Spiking Neural Networks
Dec 17, 2023



Abstract:The Spiking Neural Network (SNN), as one of the biologically inspired neural network infrastructures, has drawn increasing attention recently. It adopts binary spike activations to transmit information, thus the multiplications of activations and weights can be substituted by additions, which brings high energy efficiency. However, in the paper, we theoretically and experimentally prove that the binary spike activation map cannot carry enough information, thus causing information loss and resulting in accuracy decreasing. To handle the problem, we propose a ternary spike neuron to transmit information. The ternary spike neuron can also enjoy the event-driven and multiplication-free operation advantages of the binary spike neuron but will boost the information capacity. Furthermore, we also embed a trainable factor in the ternary spike neuron to learn the suitable spike amplitude, thus our SNN will adopt different spike amplitudes along layers, which can better suit the phenomenon that the membrane potential distributions are different along layers. To retain the efficiency of the vanilla ternary spike, the trainable ternary spike SNN will be converted to a standard one again via a re-parameterization technique in the inference. Extensive experiments with several popular network structures over static and dynamic datasets show that the ternary spike can consistently outperform state-of-the-art methods. Our code is open-sourced at https://github.com/yfguo91/Ternary-Spike.
Membrane Potential Batch Normalization for Spiking Neural Networks
Aug 16, 2023



Abstract:As one of the energy-efficient alternatives of conventional neural networks (CNNs), spiking neural networks (SNNs) have gained more and more interest recently. To train the deep models, some effective batch normalization (BN) techniques are proposed in SNNs. All these BNs are suggested to be used after the convolution layer as usually doing in CNNs. However, the spiking neuron is much more complex with the spatio-temporal dynamics. The regulated data flow after the BN layer will be disturbed again by the membrane potential updating operation before the firing function, i.e., the nonlinear activation. Therefore, we advocate adding another BN layer before the firing function to normalize the membrane potential again, called MPBN. To eliminate the induced time cost of MPBN, we also propose a training-inference-decoupled re-parameterization technique to fold the trained MPBN into the firing threshold. With the re-parameterization technique, the MPBN will not introduce any extra time burden in the inference. Furthermore, the MPBN can also adopt the element-wised form, while these BNs after the convolution layer can only use the channel-wised form. Experimental results show that the proposed MPBN performs well on both popular non-spiking static and neuromorphic datasets. Our code is open-sourced at \href{https://github.com/yfguo91/MPBN}{MPBN}.
RMP-Loss: Regularizing Membrane Potential Distribution for Spiking Neural Networks
Aug 13, 2023Abstract:Spiking Neural Networks (SNNs) as one of the biology-inspired models have received much attention recently. It can significantly reduce energy consumption since they quantize the real-valued membrane potentials to 0/1 spikes to transmit information thus the multiplications of activations and weights can be replaced by additions when implemented on hardware. However, this quantization mechanism will inevitably introduce quantization error, thus causing catastrophic information loss. To address the quantization error problem, we propose a regularizing membrane potential loss (RMP-Loss) to adjust the distribution which is directly related to quantization error to a range close to the spikes. Our method is extremely simple to implement and straightforward to train an SNN. Furthermore, it is shown to consistently outperform previous state-of-the-art methods over different network architectures and datasets.
Reducing Information Loss for Spiking Neural Networks
Jul 10, 2023



Abstract:The Spiking Neural Network (SNN) has attracted more and more attention recently. It adopts binary spike signals to transmit information. Benefitting from the information passing paradigm of SNNs, the multiplications of activations and weights can be replaced by additions, which are more energy-efficient. However, its ``Hard Reset" mechanism for the firing activity would ignore the difference among membrane potentials when the membrane potential is above the firing threshold, causing information loss. Meanwhile, quantifying the membrane potential to 0/1 spikes at the firing instants will inevitably introduce the quantization error thus bringing about information loss too. To address these problems, we propose to use the ``Soft Reset" mechanism for the supervised training-based SNNs, which will drive the membrane potential to a dynamic reset potential according to its magnitude, and Membrane Potential Rectifier (MPR) to reduce the quantization error via redistributing the membrane potential to a range close to the spikes. Results show that the SNNs with the ``Soft Reset" mechanism and MPR outperform their vanilla counterparts on both static and dynamic datasets.
Direct Learning-Based Deep Spiking Neural Networks: A Review
Jun 04, 2023Abstract:The spiking neural network (SNN), as a promising brain-inspired computational model with binary spike information transmission mechanism, rich spatially-temporal dynamics, and event-driven characteristics, has received extensive attention. However, its intricately discontinuous spike mechanism brings difficulty to the optimization of the deep SNN. Since the surrogate gradient method can greatly mitigate the optimization difficulty and shows great potential in directly training deep SNNs, a variety of direct learning-based deep SNN works have been proposed and achieved satisfying progress in recent years. In this paper, we present a comprehensive survey of these direct learning-based deep SNN works, mainly categorized into accuracy improvement methods, efficiency improvement methods, and temporal dynamics utilization methods. In addition, we also divide these categorizations into finer granularities further to better organize and introduce them. Finally, the challenges and trends that may be faced in future research are prospected.
Joint A-SNN: Joint Training of Artificial and Spiking Neural Networks via Self-Distillation and Weight Factorization
May 03, 2023



Abstract:Emerged as a biology-inspired method, Spiking Neural Networks (SNNs) mimic the spiking nature of brain neurons and have received lots of research attention. SNNs deal with binary spikes as their activation and therefore derive extreme energy efficiency on hardware. However, it also leads to an intrinsic obstacle that training SNNs from scratch requires a re-definition of the firing function for computing gradient. Artificial Neural Networks (ANNs), however, are fully differentiable to be trained with gradient descent. In this paper, we propose a joint training framework of ANN and SNN, in which the ANN can guide the SNN's optimization. This joint framework contains two parts: First, the knowledge inside ANN is distilled to SNN by using multiple branches from the networks. Second, we restrict the parameters of ANN and SNN, where they share partial parameters and learn different singular weights. Extensive experiments over several widely used network structures show that our method consistently outperforms many other state-of-the-art training methods. For example, on the CIFAR100 classification task, the spiking ResNet-18 model trained by our method can reach to 77.39% top-1 accuracy with only 4 time steps.
Real Spike: Learning Real-valued Spikes for Spiking Neural Networks
Oct 13, 2022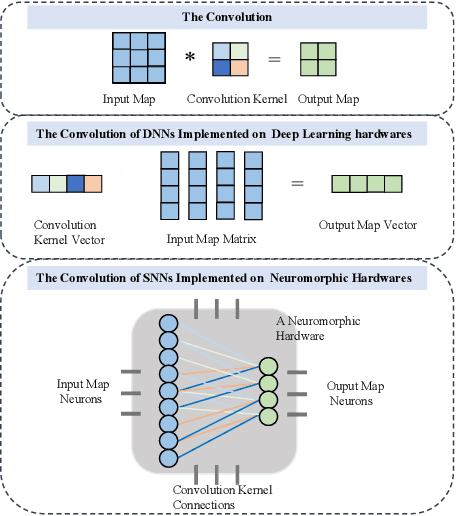
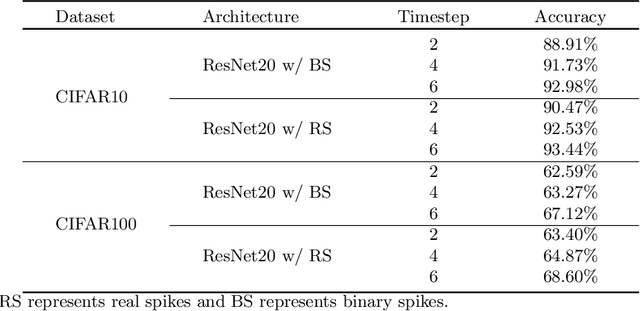
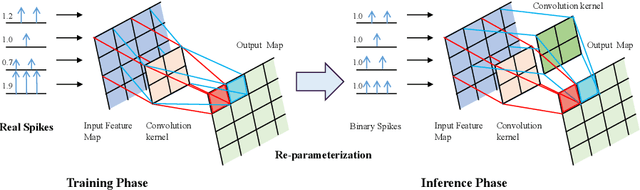
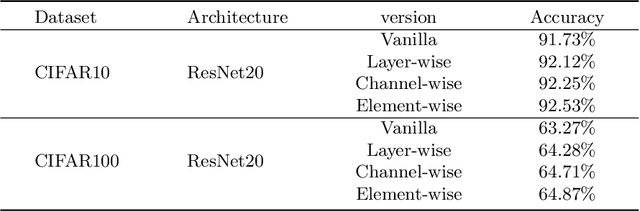
Abstract:Brain-inspired spiking neural networks (SNNs) have recently drawn more and more attention due to their event-driven and energy-efficient characteristics. The integration of storage and computation paradigm on neuromorphic hardwares makes SNNs much different from Deep Neural Networks (DNNs). In this paper, we argue that SNNs may not benefit from the weight-sharing mechanism, which can effectively reduce parameters and improve inference efficiency in DNNs, in some hardwares, and assume that an SNN with unshared convolution kernels could perform better. Motivated by this assumption, a training-inference decoupling method for SNNs named as Real Spike is proposed, which not only enjoys both unshared convolution kernels and binary spikes in inference-time but also maintains both shared convolution kernels and Real-valued Spikes during training. This decoupling mechanism of SNN is realized by a re-parameterization technique. Furthermore, based on the training-inference-decoupled idea, a series of different forms for implementing Real Spike on different levels are presented, which also enjoy shared convolutions in the inference and are friendly to both neuromorphic and non-neuromorphic hardware platforms. A theoretical proof is given to clarify that the Real Spike-based SNN network is superior to its vanilla counterpart. Experimental results show that all different Real Spike versions can consistently improve the SNN performance. Moreover, the proposed method outperforms the state-of-the-art models on both non-spiking static and neuromorphic datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge