Xiuli Liu
Britton Chance Center for Biomedical Photonics, Wuhan National Laboratory for Optoelectronics-Huazhong University of Science and Technology, China
ParamNet: A Parameter-variable Network for Fast Stain Normalization
May 11, 2023



Abstract:In practice, digital pathology images are often affected by various factors, resulting in very large differences in color and brightness. Stain normalization can effectively reduce the differences in color and brightness of digital pathology images, thus improving the performance of computer-aided diagnostic systems. Conventional stain normalization methods rely on one or several reference images, but one or several images are difficult to represent the entire dataset. Although learning-based stain normalization methods are a general approach, they use complex deep networks, which not only greatly reduce computational efficiency, but also risk introducing artifacts. StainNet is a fast and robust stain normalization network, but it has not a sufficient capability for complex stain normalization due to its too simple network structure. In this study, we proposed a parameter-variable stain normalization network, ParamNet. ParamNet contains a parameter prediction sub-network and a color mapping sub-network, where the parameter prediction sub-network can automatically determine the appropriate parameters for the color mapping sub-network according to each input image. The feature of parameter variable ensures that our network has a sufficient capability for various stain normalization tasks. The color mapping sub-network is a fully 1x1 convolutional network with a total of 59 variable parameters, which allows our network to be extremely computationally efficient and does not introduce artifacts. The results on cytopathology and histopathology datasets show that our ParamNet outperforms state-of-the-art methods and can effectively improve the generalization of classifiers on pathology diagnosis tasks. The code has been available at https://github.com/khtao/ParamNet.
Cervical Glandular Cell Detection from Whole Slide Image with Out-Of-Distribution Data
Jun 01, 2022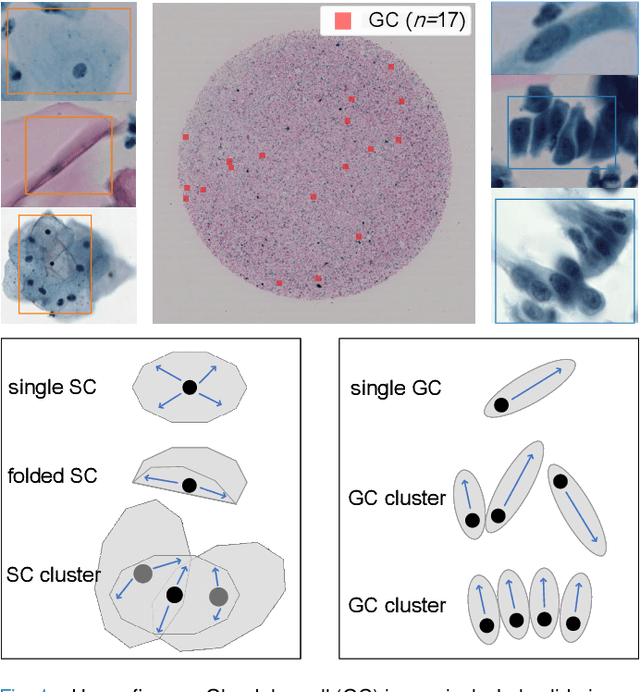
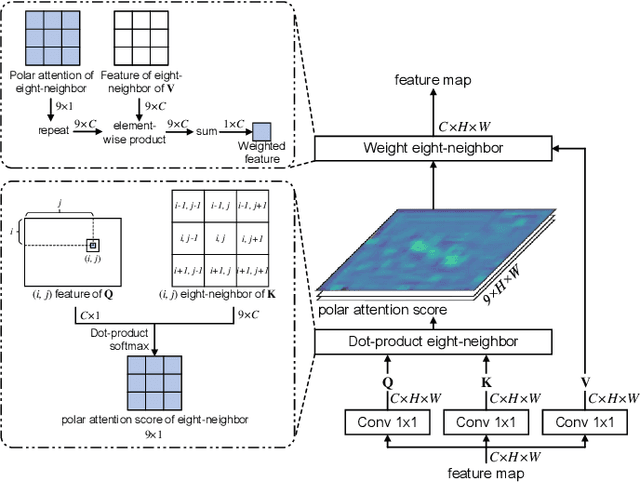
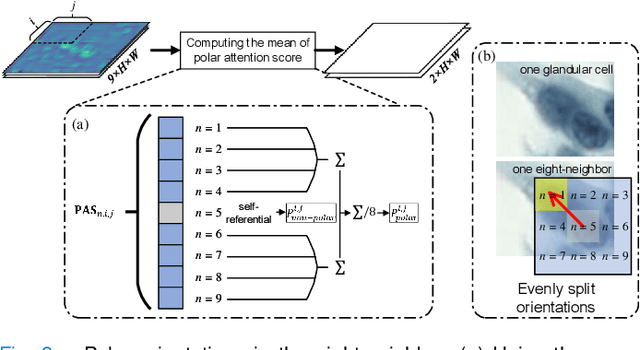
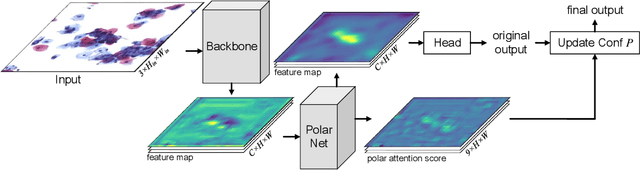
Abstract:Cervical glandular cell (GC) detection is a key step in computer-aided diagnosis for cervical adenocarcinomas screening. It is challenging to accurately recognize GCs in cervical smears in which squamous cells are the major. Widely existing Out-Of-Distribution (OOD) data in the entire smear leads decreasing reliability of machine learning system for GC detection. Although, the State-Of-The-Art (SOTA) deep learning model can outperform pathologists in preselected regions of interest, the mass False Positive (FP) prediction with high probability is still unsolved when facing such gigapixel whole slide image. This paper proposed a novel PolarNet based on the morphological prior knowledge of GC trying to solve the FP problem via a self-attention mechanism in eight-neighbor. It estimates the polar orientation of nucleus of GC. As a plugin module, PolarNet can guide the deep feature and predicted confidence of general object detection models. In experiments, we discovered that general models based on four different frameworks can reject FP in small image set and increase the mean of average precision (mAP) by $\text{0.007}\sim\text{0.015}$ in average, where the highest exceeds the recent cervical cell detection model 0.037. By plugging PolarNet, the deployed C++ program improved by 8.8\% on accuracy of top-20 GC detection from external WSIs, while sacrificing 14.4 s of computational time. Code is available in https://github.com/Chrisa142857/PolarNet-GCdet
An Efficient Cervical Whole Slide Image Analysis Framework Based on Multi-scale Semantic and Spatial Deep Features
Jul 03, 2021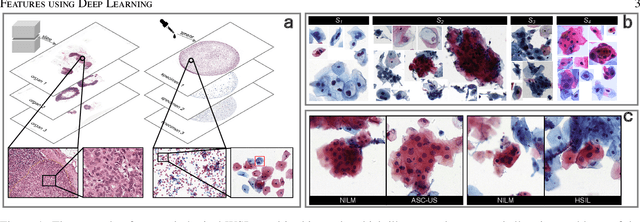
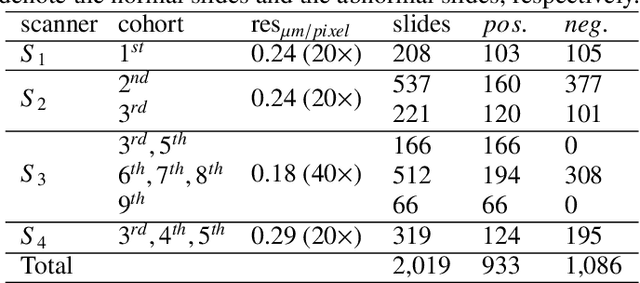
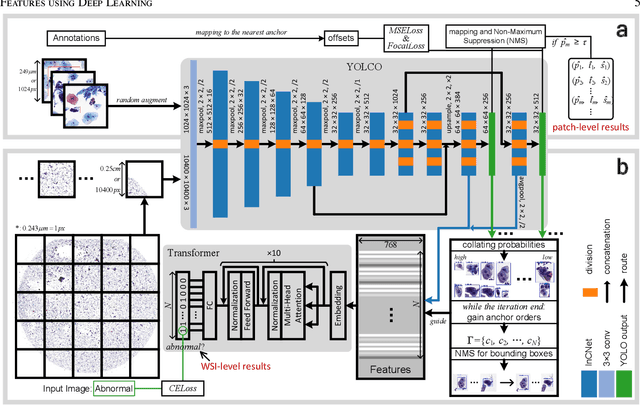
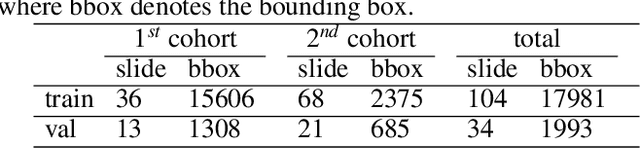
Abstract:Digital gigapixel whole slide image (WSI) is widely used in clinical diagnosis, and automated WSI analysis is key for computer-aided diagnosis. Currently, analyzing the integrated descriptor of probabilities or feature maps from massive local patches encoded by ResNet classifier is the main manner for WSI-level prediction. Feature representations of the sparse and tiny lesion cells in cervical slides, however, are still challengeable for the under-promoted upstream encoders, while the unused spatial representations of cervical cells are the available features to supply the semantics analysis. As well as patches sampling with overlap and repetitive processing incur the inefficiency and the unpredictable side effect. This study designs a novel inline connection network (InCNet) by enriching the multi-scale connectivity to build the lightweight model named You Only Look Cytopathology Once (YOLCO) with the additional supervision of spatial information. The proposed model allows the input size enlarged to megapixel that can stitch the WSI without any overlap by the average repeats decreased from $10^3\sim10^4$ to $10^1\sim10^2$ for collecting features and predictions at two scales. Based on Transformer for classifying the integrated multi-scale multi-task features, the experimental results appear $0.872$ AUC score better and $2.51\times$ faster than the best conventional method in WSI classification on multicohort datasets of 2,019 slides from four scanning devices.
StainNet: a fast and robust stain normalization network
Jan 16, 2021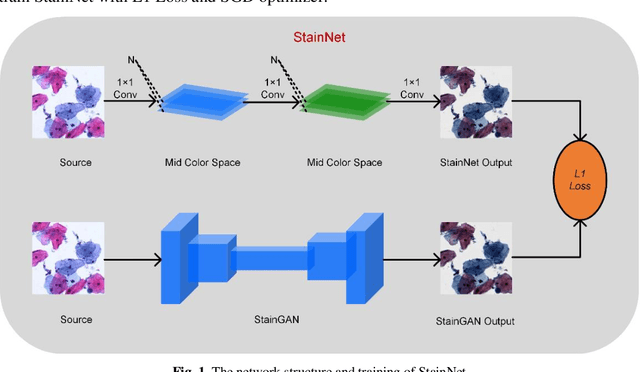
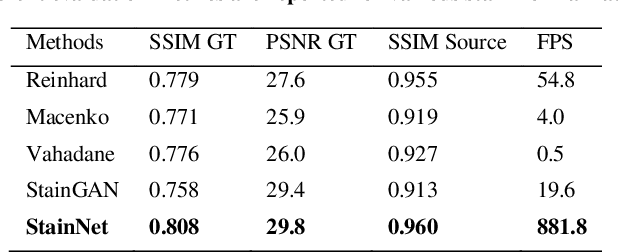
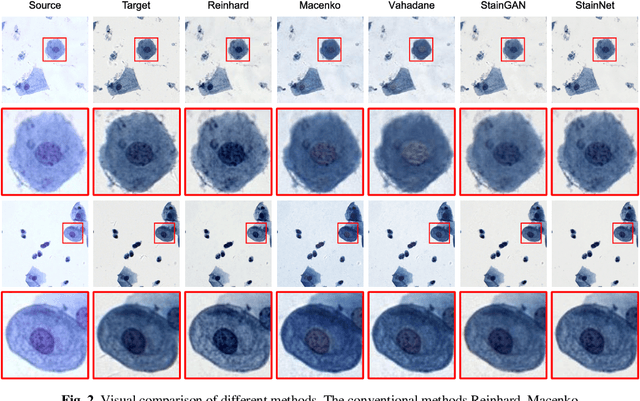
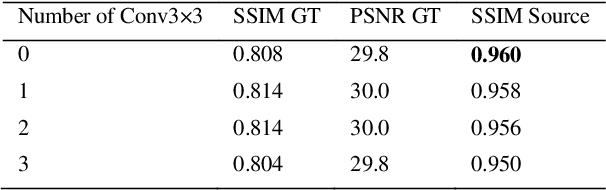
Abstract:Due to a variety of factors, pathological images have large color variabilities, which hamper the performance of computer-aided diagnosis (CAD) systems. Stain normalization has been used to reduce the color variability and increase the accuracy of CAD systems. Among them, the conventional methods perform stain normalization on a pixel-by-pixel basis, but estimate stain parameters just relying on one single reference image and thus would incur some inaccurate normalization results. As for the current deep learning-based methods, it can automatically extract the color distribution and need not pick a representative reference image. While the deep learning-based methods have a complex structure with millions of parameters, and a relatively low computational efficiency and a risk to introduce artifacts. In this paper, a fast and robust stain normalization network with only 1.28K parameters named StainNet is proposed. StainNet can learn the color mapping relationship from a whole dataset and adjust the color value in a pixel-to-pixel manner. The proposed method performs well in stain normalization and achieves a better accuracy and image quality. Application results show the cervical cytology classification achieved a higher accuracy when after stain normalization of StainNet.
Reconstruct high-resolution multi-focal plane images from a single 2D wide field image
Sep 21, 2020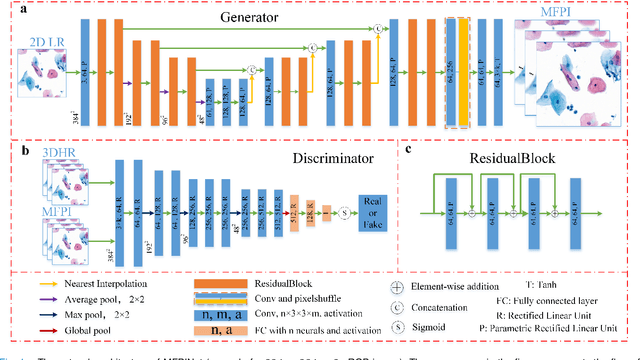
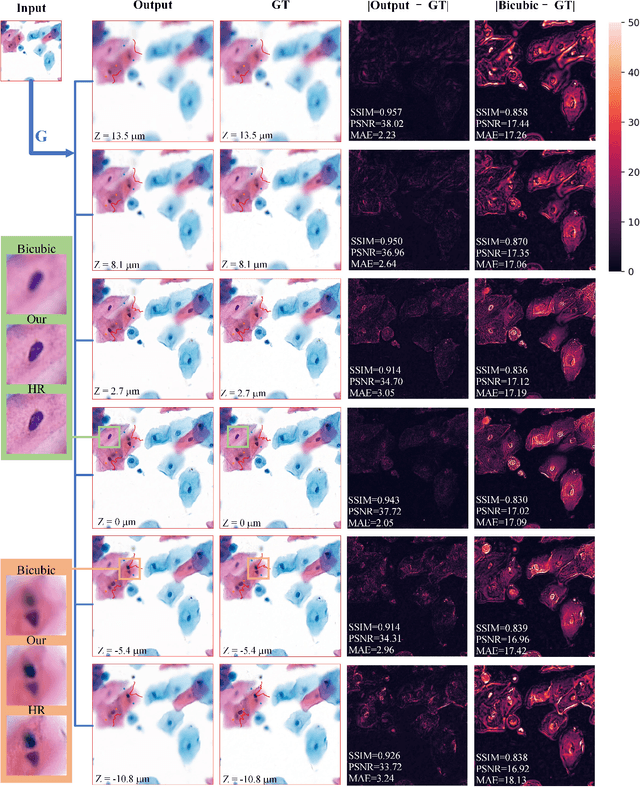
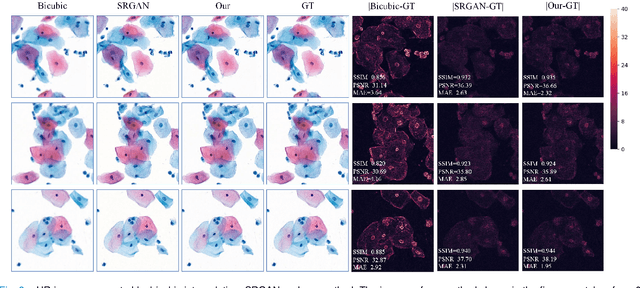
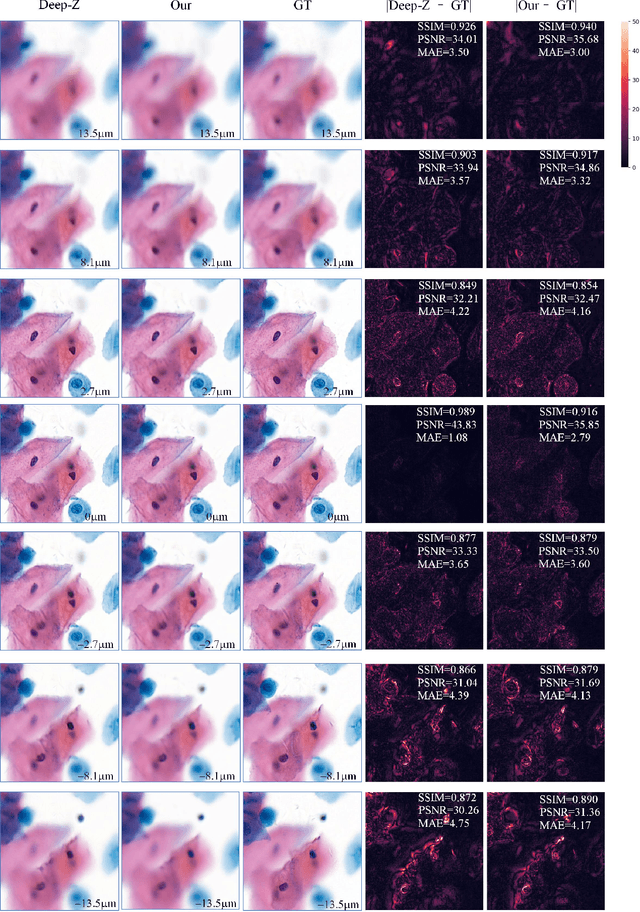
Abstract:High-resolution 3D medical images are important for analysis and diagnosis, but axial scanning to acquire them is very time-consuming. In this paper, we propose a fast end-to-end multi-focal plane imaging network (MFPINet) to reconstruct high-resolution multi-focal plane images from a single 2D low-resolution wild filed image without relying on scanning. To acquire realistic MFP images fast, the proposed MFPINet adopts generative adversarial network framework and the strategies of post-sampling and refocusing all focal planes at one time. We conduct a series experiments on cytology microscopy images and demonstrate that MFPINet performs well on both axial refocusing and horizontal super resolution. Furthermore, MFPINet is approximately 24 times faster than current refocusing methods for reconstructing the same volume images. The proposed method has the potential to greatly increase the speed of high-resolution 3D imaging and expand the application of low-resolution wide-field images.
FFusionCGAN: An end-to-end fusion method for few-focus images using conditional GAN in cytopathological digital slides
Jan 03, 2020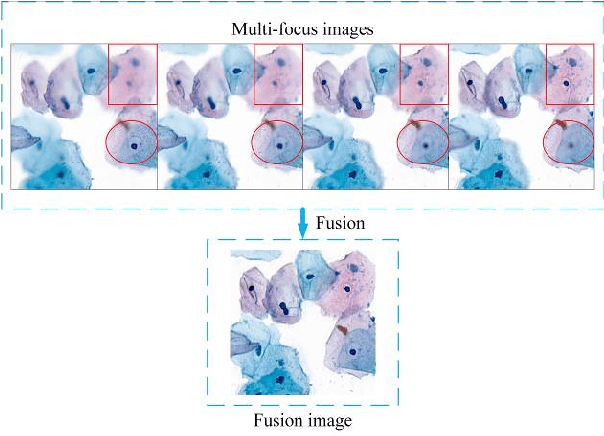
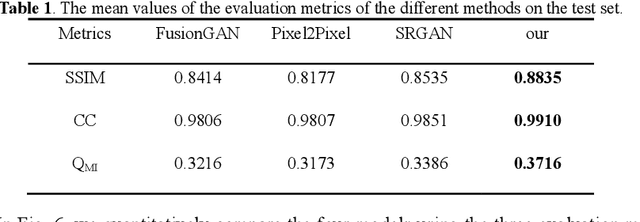
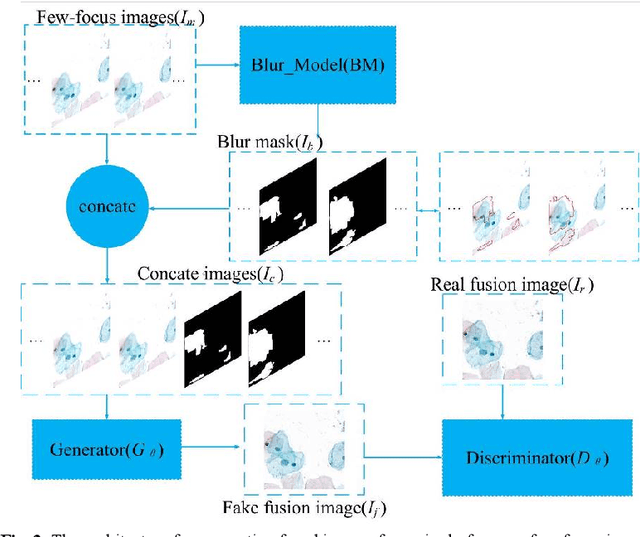
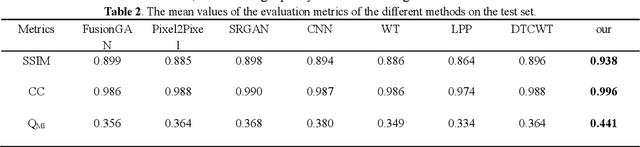
Abstract:Multi-focus image fusion technologies compress different focus depth images into an image in which most objects are in focus. However, although existing image fusion techniques, including traditional algorithms and deep learning-based algorithms, can generate high-quality fused images, they need multiple images with different focus depths in the same field of view. This criterion may not be met in some cases where time efficiency is required or the hardware is insufficient. The problem is especially prominent in large-size whole slide images. This paper focused on the multi-focus image fusion of cytopathological digital slide images, and proposed a novel method for generating fused images from single-focus or few-focus images based on conditional generative adversarial network (GAN). Through the adversarial learning of the generator and discriminator, the method is capable of generating fused images with clear textures and large depth of field. Combined with the characteristics of cytopathological images, this paper designs a new generator architecture combining U-Net and DenseBlock, which can effectively improve the network's receptive field and comprehensively encode image features. Meanwhile, this paper develops a semantic segmentation network that identifies the blurred regions in cytopathological images. By integrating the network into the generative model, the quality of the generated fused images is effectively improved. Our method can generate fused images from only single-focus or few-focus images, thereby avoiding the problem of collecting multiple images of different focus depths with increased time and hardware costs. Furthermore, our model is designed to learn the direct mapping of input source images to fused images without the need to manually design complex activity level measurements and fusion rules as in traditional methods.
Multi-stage domain adversarial style reconstruction for cytopathological image stain normalization
Sep 11, 2019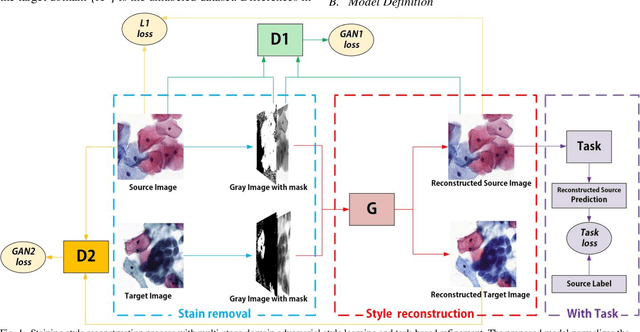
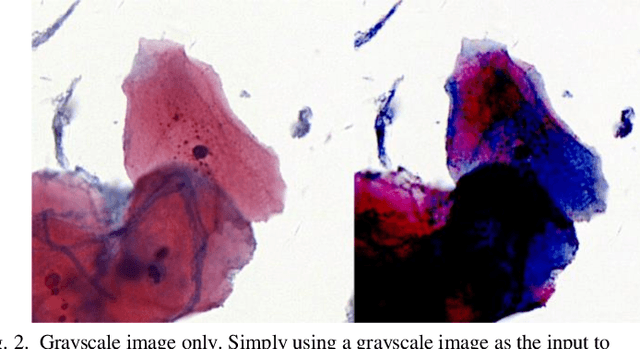
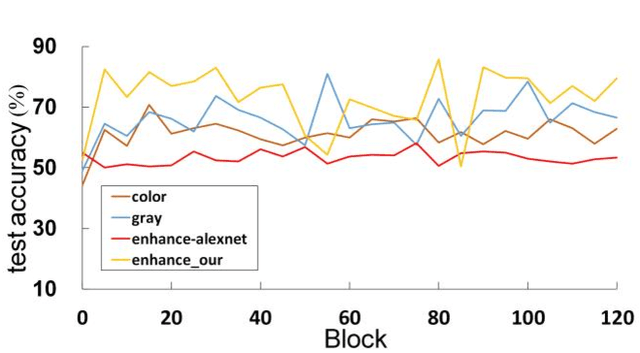
Abstract:The different stain styles of cytopathological images have a negative effect on the generalization ability of automated image analysis algorithms. This article proposes a new framework that normalizes the stain style for cytopathological images through a stain removal module and a multi-stage domain adversarial style reconstruction module. We convert colorful images into grayscale images with a color-encoding mask. Using the mask, reconstructed images retain their basic color without red and blue mixing, which is important for cytopathological image interpretation. The style reconstruction module consists of per-pixel regression with intradomain adversarial learning, inter-domain adversarial learning, and optional task-based refining. Per-pixel regression with intradomain adversarial learning establishes the generative network from the decolorized input to the reconstructed output. The interdomain adversarial learning further reduces the difference in stain style. The generation network can be optimized by combining it with the task network. Experimental results show that the proposed techniques help to optimize the generation network. The average accuracy increases from 75.41% to 84.79% after the intra-domain adversarial learning, and to 87.00% after interdomain adversarial learning. Under the guidance of the task network, the average accuracy rate reaches 89.58%. The proposed method achieves unsupervised stain normalization of cytopathological images, while preserving the cell structure, texture structure, and cell color properties of the image. This method overcomes the problem of generalizing the task models between different stain styles of cytopathological images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge