Xinzhuo Jiang
Columbia University Irving Medical Center
One Loss to Rule Them All: Marked Time-to-Event for Structured EHR Foundation Models
Jan 31, 2026Abstract:Clinical events captured in Electronic Health Records (EHR) are irregularly sampled and may consist of a mixture of discrete events and numerical measurements, such as laboratory values or treatment dosages. The sequential nature of EHR, analogous to natural language, has motivated the use of next-token prediction to train prior EHR Foundation Models (FMs) over events. However, this training fails to capture the full structure of EHR. We propose ORA, a marked time-to-event pretraining objective that jointly models event timing and associated measurements. Across multiple datasets, downstream tasks, and model architectures, this objective consistently yields more generalizable representations than next-token prediction and pretraining losses that ignore continuous measurements. Importantly, the proposed objective yields improvements beyond traditional classification evaluation, including better regression and time-to-event prediction. Beyond introducing a new family of FMs, our results suggest a broader takeaway: pretraining objectives that account for EHR structure are critical for expanding downstream capabilities and generalizability
CEHR-GPT: A Scalable Multi-Task Foundation Model for Electronic Health Records
Sep 03, 2025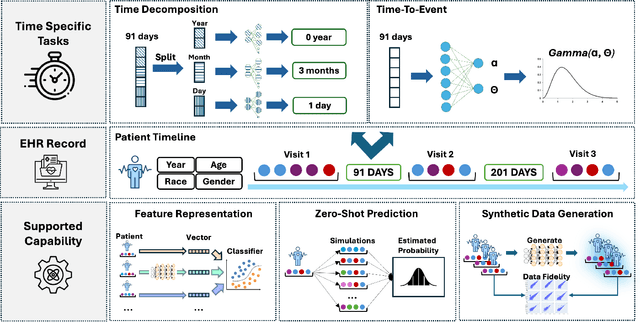

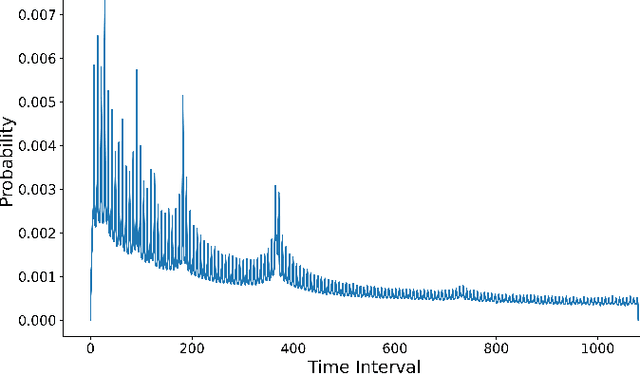

Abstract:Electronic Health Records (EHRs) provide a rich, longitudinal view of patient health and hold significant potential for advancing clinical decision support, risk prediction, and data-driven healthcare research. However, most artificial intelligence (AI) models for EHRs are designed for narrow, single-purpose tasks, limiting their generalizability and utility in real-world settings. Here, we present CEHR-GPT, a general-purpose foundation model for EHR data that unifies three essential capabilities - feature representation, zero-shot prediction, and synthetic data generation - within a single architecture. To support temporal reasoning over clinical sequences, \cehrgpt{} incorporates a novel time-token-based learning framework that explicitly encodes patients' dynamic timelines into the model structure. CEHR-GPT demonstrates strong performance across all three tasks and generalizes effectively to external datasets through vocabulary expansion and fine-tuning. Its versatility enables rapid model development, cohort discovery, and patient outcome forecasting without the need for task-specific retraining.
FoMoH: A clinically meaningful foundation model evaluation for structured electronic health records
May 22, 2025Abstract:Foundation models hold significant promise in healthcare, given their capacity to extract meaningful representations independent of downstream tasks. This property has enabled state-of-the-art performance across several clinical applications trained on structured electronic health record (EHR) data, even in settings with limited labeled data, a prevalent challenge in healthcare. However, there is little consensus on these models' potential for clinical utility due to the lack of desiderata of comprehensive and meaningful tasks and sufficiently diverse evaluations to characterize the benefit over conventional supervised learning. To address this gap, we propose a suite of clinically meaningful tasks spanning patient outcomes, early prediction of acute and chronic conditions, including desiderata for robust evaluations. We evaluate state-of-the-art foundation models on EHR data consisting of 5 million patients from Columbia University Irving Medical Center (CUMC), a large urban academic medical center in New York City, across 14 clinically relevant tasks. We measure overall accuracy, calibration, and subpopulation performance to surface tradeoffs based on the choice of pre-training, tokenization, and data representation strategies. Our study aims to advance the empirical evaluation of structured EHR foundation models and guide the development of future healthcare foundation models.
CEHR-GPT: Generating Electronic Health Records with Chronological Patient Timelines
Feb 06, 2024



Abstract:Synthetic Electronic Health Records (EHR) have emerged as a pivotal tool in advancing healthcare applications and machine learning models, particularly for researchers without direct access to healthcare data. Although existing methods, like rule-based approaches and generative adversarial networks (GANs), generate synthetic data that resembles real-world EHR data, these methods often use a tabular format, disregarding temporal dependencies in patient histories and limiting data replication. Recently, there has been a growing interest in leveraging Generative Pre-trained Transformers (GPT) for EHR data. This enables applications like disease progression analysis, population estimation, counterfactual reasoning, and synthetic data generation. In this work, we focus on synthetic data generation and demonstrate the capability of training a GPT model using a particular patient representation derived from CEHR-BERT, enabling us to generate patient sequences that can be seamlessly converted to the Observational Medical Outcomes Partnership (OMOP) data format.
CEHR-BERT: Incorporating temporal information from structured EHR data to improve prediction tasks
Nov 10, 2021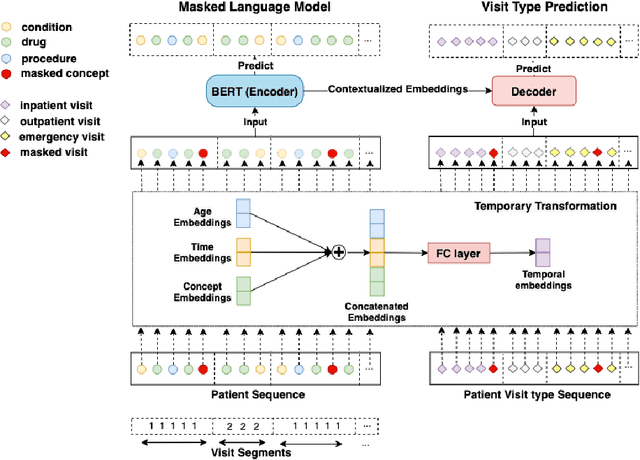
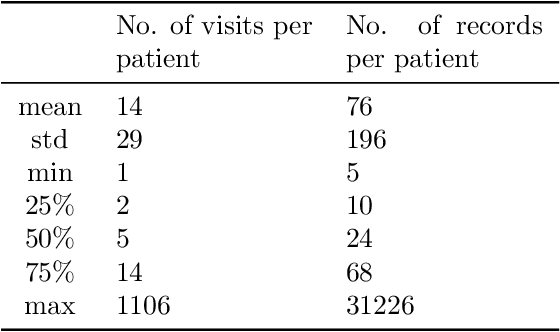
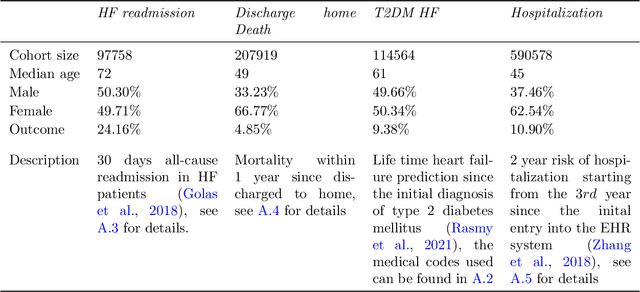
Abstract:Embedding algorithms are increasingly used to represent clinical concepts in healthcare for improving machine learning tasks such as clinical phenotyping and disease prediction. Recent studies have adapted state-of-the-art bidirectional encoder representations from transformers (BERT) architecture to structured electronic health records (EHR) data for the generation of contextualized concept embeddings, yet do not fully incorporate temporal data across multiple clinical domains. Therefore we developed a new BERT adaptation, CEHR-BERT, to incorporate temporal information using a hybrid approach by augmenting the input to BERT using artificial time tokens, incorporating time, age, and concept embeddings, and introducing a new second learning objective for visit type. CEHR-BERT was trained on a subset of Columbia University Irving Medical Center-York Presbyterian Hospital's clinical data, which includes 2.4M patients, spanning over three decades, and tested using 4-fold cross-validation on the following prediction tasks: hospitalization, death, new heart failure (HF) diagnosis, and HF readmission. Our experiments show that CEHR-BERT outperformed existing state-of-the-art clinical BERT adaptations and baseline models across all 4 prediction tasks in both ROC-AUC and PR-AUC. CEHR-BERT also demonstrated strong transfer learning capability, as our model trained on only 5% of data outperformed comparison models trained on the entire data set. Ablation studies to better understand the contribution of each time component showed incremental gains with every element, suggesting that CEHR-BERT's incorporation of artificial time tokens, time and age embeddings with concept embeddings, and the addition of the second learning objective represents a promising approach for future BERT-based clinical embeddings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge