Xiaoyi Jiang
A Streaming Volumetric Image Generation Framework for Development and Evaluation of Out-of-Core Methods
Dec 18, 2021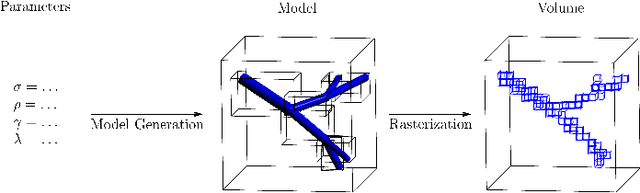
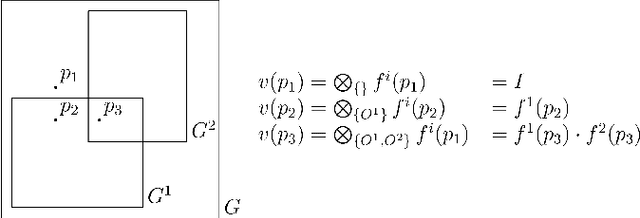
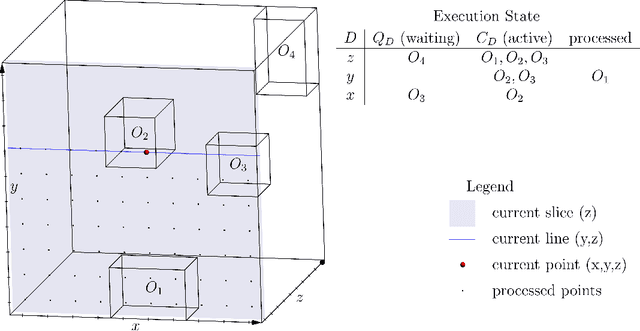
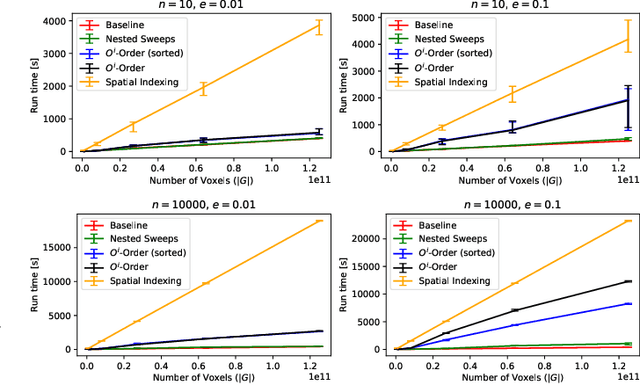
Abstract:Advances in 3D imaging technology in recent years have allowed for increasingly high resolution volumetric images of large specimen. The resulting datasets of hundreds of Gigabytes in size call for new scalable and memory efficient approaches in the field of image processing, where some progress has been made already. At the same time, quantitative evaluation of these new methods is difficult both in terms of the availability of specific data sizes and in the generation of associated ground truth data. In this paper we present an algorithmic framework that can be used to efficiently generate test (and ground truth) volume data, optionally even in a streaming fashion. As the proposed nested sweeps algorithm is fast, it can be used to generate test data on demand. We analyze the asymptotic run time of the presented algorithm and compare it experimentally to alternative approaches as well as a hypothetical best-case baseline method. In a case study, the framework is applied to the popular VascuSynth software for vascular image generation, making it capable of efficiently producing larger-than-main memory volumes which is demonstrated by generating a trillion voxel (1TB) image. Implementations of the presented framework are available online in the form of the modified version of Vascusynth and the code used for the experimental evaluation. In addition, the test data generation procedure has been integrated into the popular volume rendering and processing framework Voreen.
Hierarchical Random Walker Segmentation for Large Volumetric Biomedical Data
Mar 17, 2021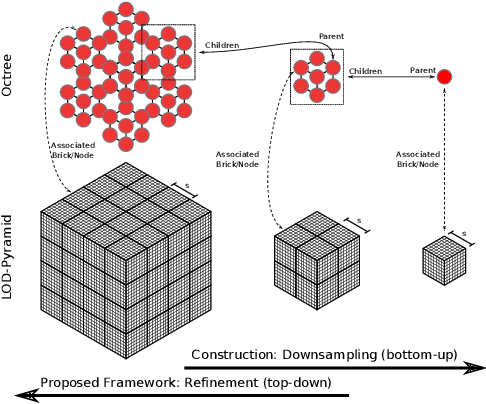
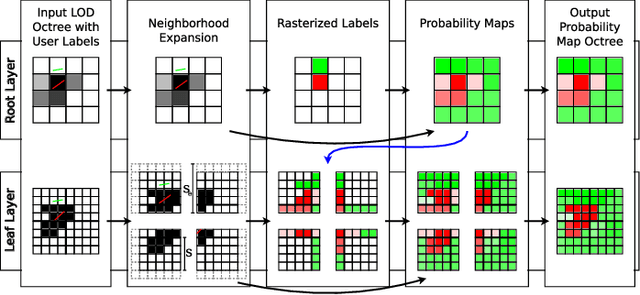
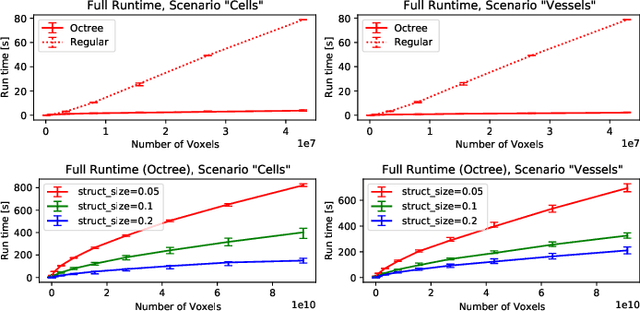
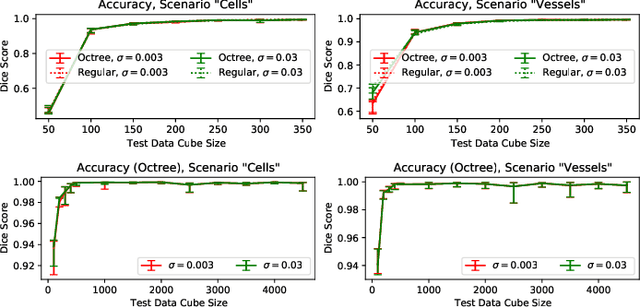
Abstract:The random walker method for image segmentation is a popular tool for semi-automatic image segmentation, especially in the biomedical field. However, its linear asymptotic run time and memory requirements make application to 3D datasets of increasing sizes impractical. We propose a hierarchical framework that, to the best of our knowledge, is the first attempt to overcome these restrictions for the random walker algorithm and achieves sublinear run time and constant memory complexity. The method is evaluated on synthetic data and real data from current biomedical research, where high segmentation quality is quantitatively confirmed and visually observed, respectively. The incremental (i.e., interaction update) run time is demonstrated to be in seconds on a standard PC even for volumes of hundreds of Gigabytes in size. An implementation of the presented method is publicly available in version 5.2 of the widely used volume rendering and processing software Voreen (https://www.uni-muenster.de/Voreen/).
Scalable Robust Graph and Feature Extraction for Arbitrary Vessel Networks in Large Volumetric Datasets
Feb 05, 2021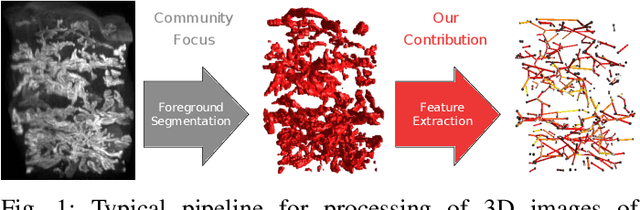
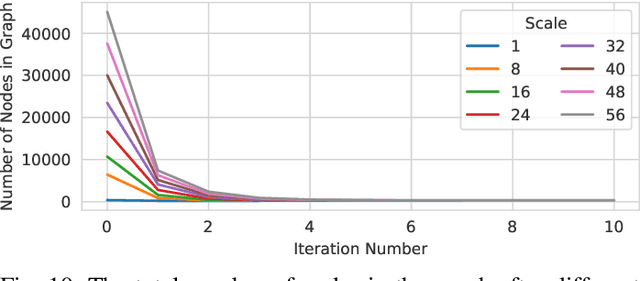
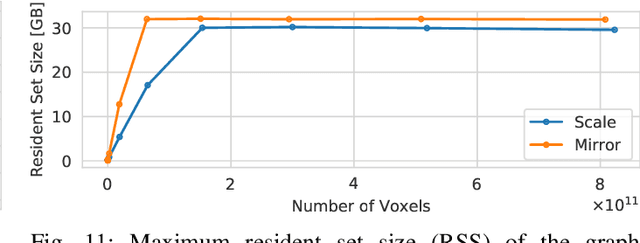
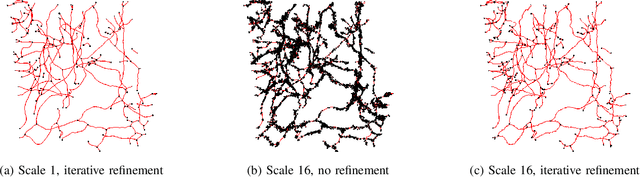
Abstract:Recent advances in 3D imaging technologies provide novel insights to researchers and reveal finer and more detail of examined specimen, especially in the biomedical domain, but also impose huge challenges regarding scalability for automated analysis algorithms due to rapidly increasing dataset sizes. In particular, existing research towards automated vessel network analysis does not consider memory requirements of proposed algorithms and often generates a large number of spurious branches for structures consisting of many voxels. Additionally, very often these algorithms have further restrictions such as the limitation to tree topologies or relying on the properties of specific image modalities. We present a scalable pipeline (in terms of computational cost, required main memory and robustness) that extracts an annotated abstract graph representation from the foreground segmentation of vessel networks of arbitrary topology and vessel shape. Only a single, dimensionless, a-priori determinable parameter is required. By careful engineering of individual pipeline stages and a novel iterative refinement scheme we are, for the first time, able to analyze the topology of volumes of roughly 1TB on commodity hardware. An implementation of the presented pipeline is publicly available in version 5.1 of the volume rendering and processing engine Voreen (https://www.uni-muenster.de/Voreen/).
The PHOTON Wizard -- Towards Educational Machine Learning Code Generators
Feb 13, 2020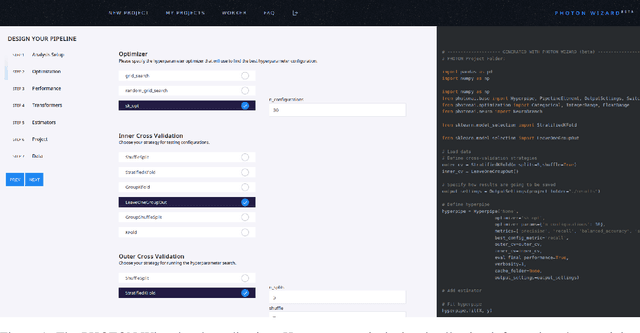
Abstract:Despite the tremendous efforts to democratize machine learning, especially in applied-science, the application is still often hampered by the lack of coding skills. As we consider programmatic understanding key to building effective and efficient machine learning solutions, we argue for a novel educational approach that builds upon the accessibility and acceptance of graphical user interfaces to convey programming skills to an applied-science target group. We outline a proof-of-concept, open-source web application, the PHOTON Wizard, which dynamically translates GUI interactions into valid source code for the Python machine learning framework PHOTON. Thereby, users possessing theoretical machine learning knowledge gain key insights into the model development workflow as well as an intuitive understanding of custom implementations. Specifically, the PHOTON Wizard integrates the concept of Educational Machine Learning Code Generators to teach users how to write code for designing, training, optimizing and evaluating custom machine learning pipelines.
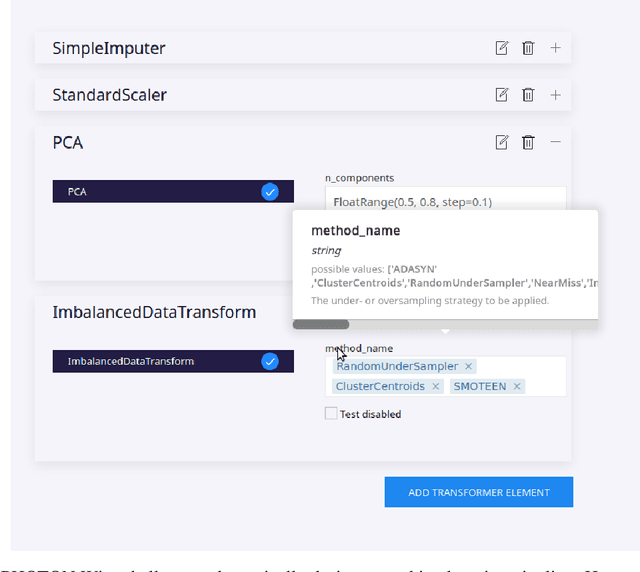
PHOTON -- A Python API for Rapid Machine Learning Model Development
Feb 13, 2020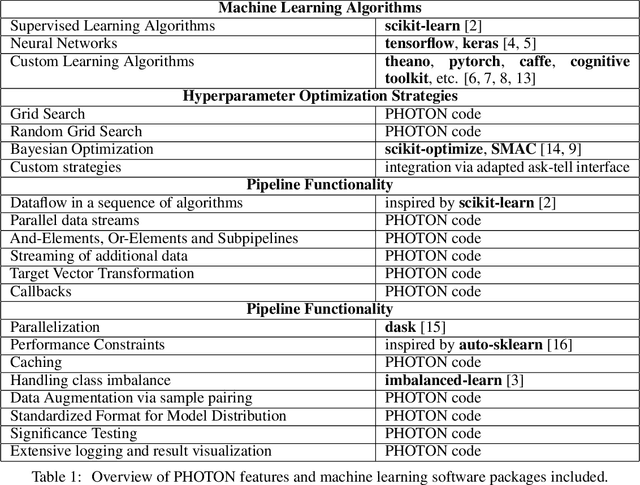
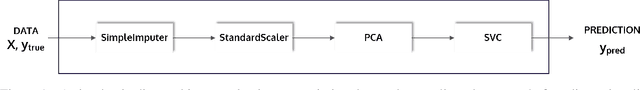
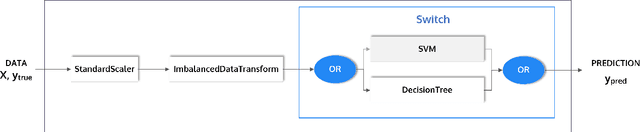
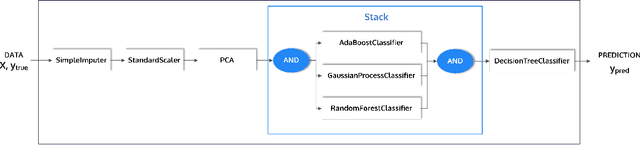
Abstract:This article describes the implementation and use of PHOTON, a high-level Python API designed to simplify and accelerate the process of machine learning model development. It enables designing both basic and advanced machine learning pipeline architectures and automatizes the repetitive training, optimization and evaluation workflow. PHOTON offers easy access to established machine learning toolboxes as well as the possibility to integrate custom algorithms and solutions for any part of the model construction and evaluation process. By adding a layer of abstraction incorporating current best practices it offers an easy-to-use, flexible approach to implementing fast, reproducible, and unbiased machine learning solutions.
Systematic Overestimation of Machine Learning Performance in Neuroimaging Studies of Depression
Dec 13, 2019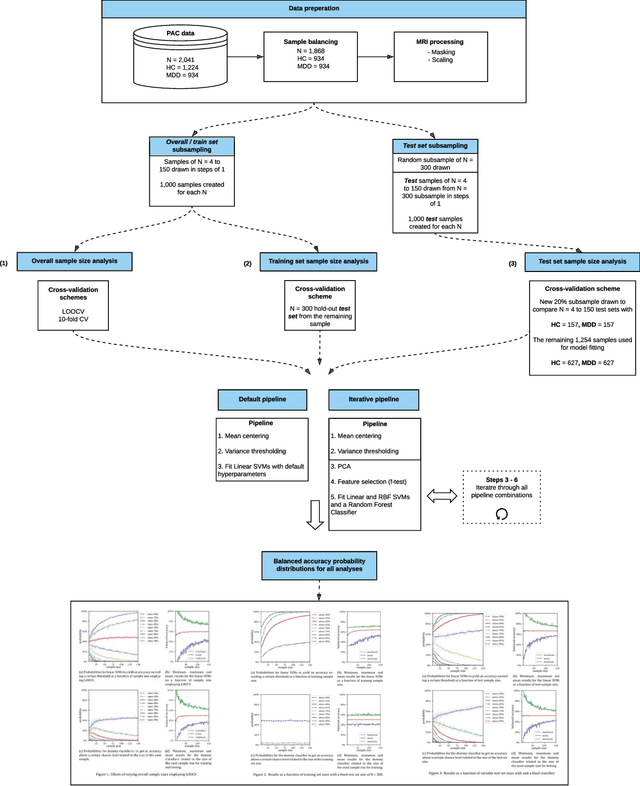
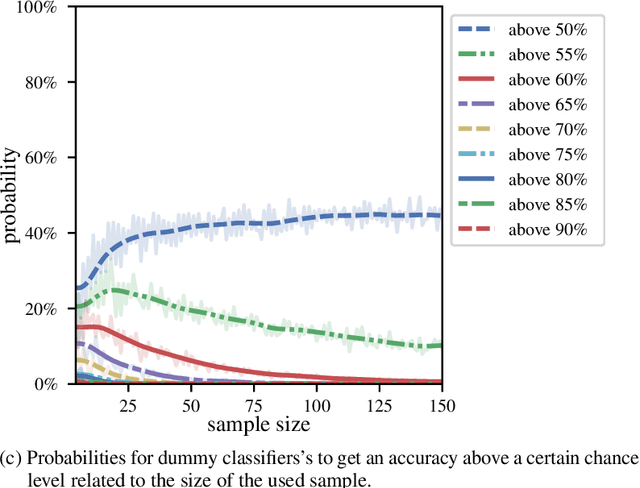
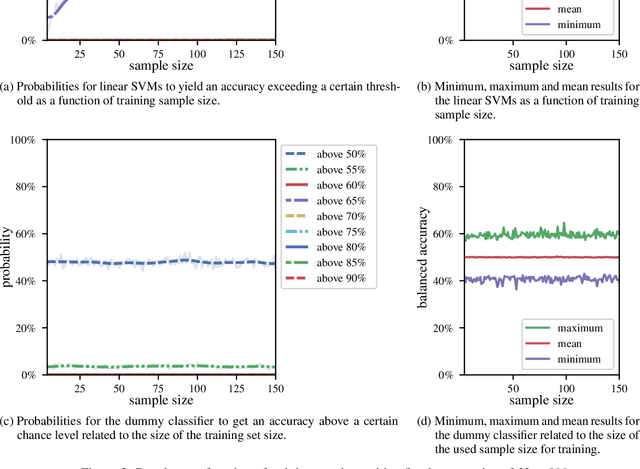
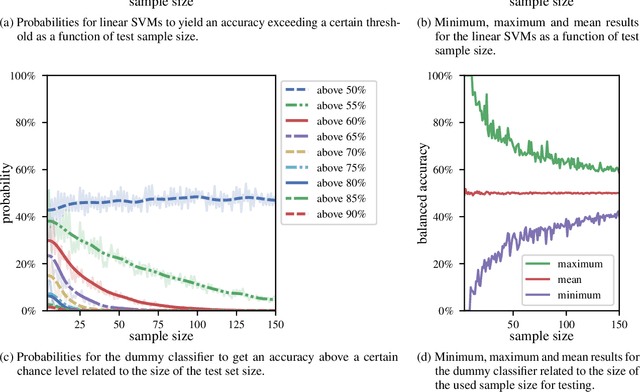
Abstract:We currently observe a disconcerting phenomenon in machine learning studies in psychiatry: While we would expect larger samples to yield better results due to the availability of more data, larger machine learning studies consistently show much weaker performance than the numerous small-scale studies. Here, we systematically investigated this effect focusing on one of the most heavily studied questions in the field, namely the classification of patients suffering from Major Depressive Disorder (MDD) and healthy controls. Drawing upon a balanced sample of $N = 1,868$ MDD patients and healthy controls from our recent international Predictive Analytics Competition (PAC), we first trained and tested a classification model on the full dataset which yielded an accuracy of 61%. Next, we mimicked the process by which researchers would draw samples of various sizes ($N=4$ to $N=150$) from the population and showed a strong risk of overestimation. Specifically, for small sample sizes ($N=20$), we observe accuracies of up to 95%. For medium sample sizes ($N=100$) accuracies up to 75% were found. Importantly, further investigation showed that sufficiently large test sets effectively protect against performance overestimation whereas larger datasets per se do not. While these results question the validity of a substantial part of the current literature, we outline the relatively low-cost remedy of larger test sets.
Biological sex classification with structural MRI data shows increased misclassification in transgender women
Nov 24, 2019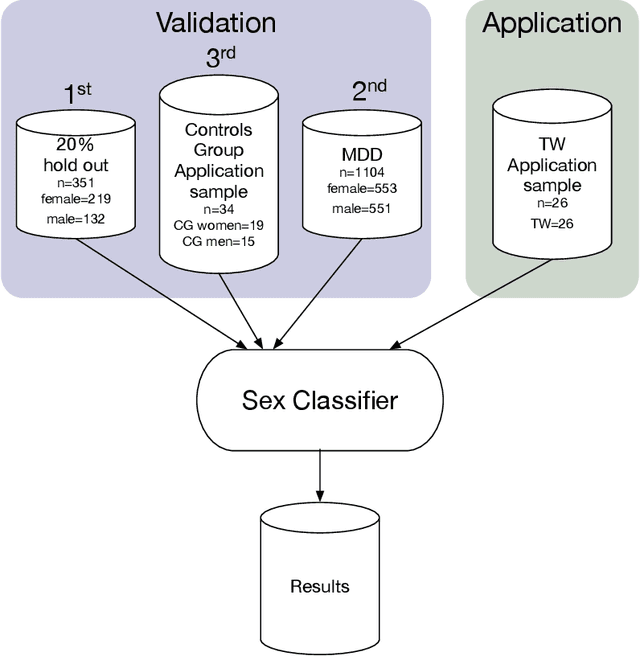
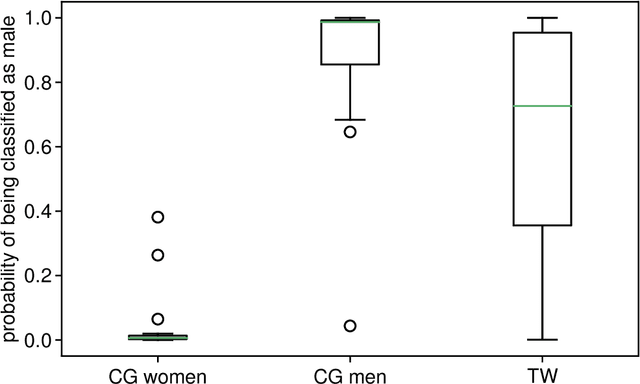
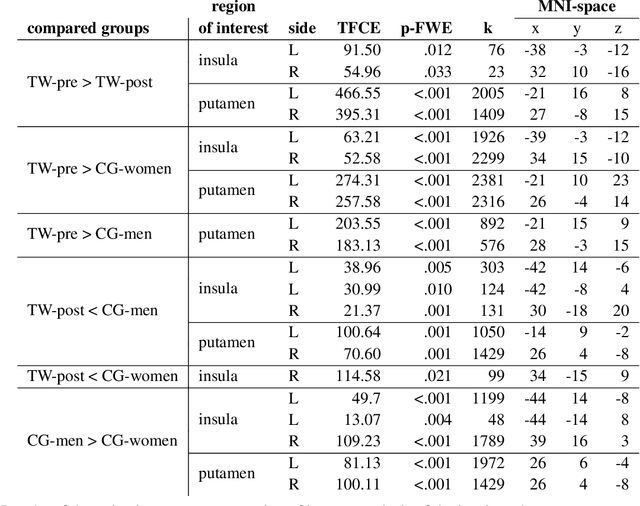
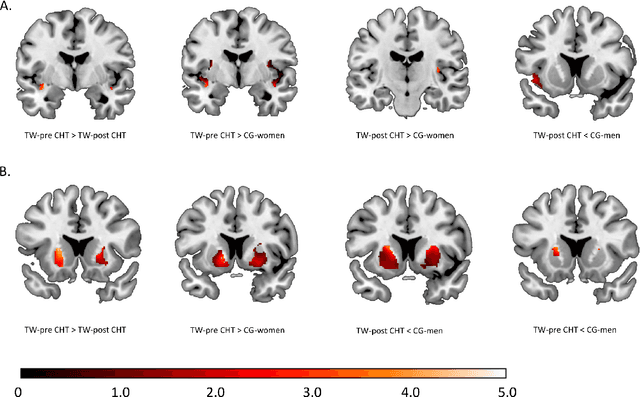
Abstract:Transgender individuals show brain structural alterations that differ from their biological sex as well as their perceived gender. To substantiate evidence that the brain structure of transgender individuals differs from male and female, we use a combined multivariate and univariate approach. Gray matter segments resulting from voxel-based morphometry preprocessing of N = 1753 cisgender (CG) healthy participants were used to train (N = 1402) and validate (20% hold-out N = 351) a support vector machine classifying the biological sex. As a second validation, we classified N = 1104 patients with depression. A third validation was performed using the matched CG sample of the transgender women (TW) application sample. Subsequently, the classifier was applied to N = 25 TW. Finally, we compared brain volumes of CG-men, women and TW pre/post treatment (CHT) in a univariate analysis controlling for sexual orientation, age and total brain volume. The application of our biological sex classifier to the transgender sample resulted in a significantly lower true positive rate (TPR-male = 56.0%). The TPR did not differ between CG-individuals with (TPR-male = 86.9%) and without depression (TPR-male = 88.5%). The univariate analysis of the transgender application sample revealed that TW pre/post treatment show brain structural differences from CG-women and CG-men in the putamen and insula, as well as the whole-brain analysis. Our results support the hypothesis that brain structure in TW differs from brain structure of their biological sex (male) as well as their perceived gender (female). This finding substantiates evidence that transgender individuals show specific brain structural alterations leading to a different pattern of brain structure than CG individuals.
Barista - a Graphical Tool for Designing and Training Deep Neural Networks
Feb 13, 2018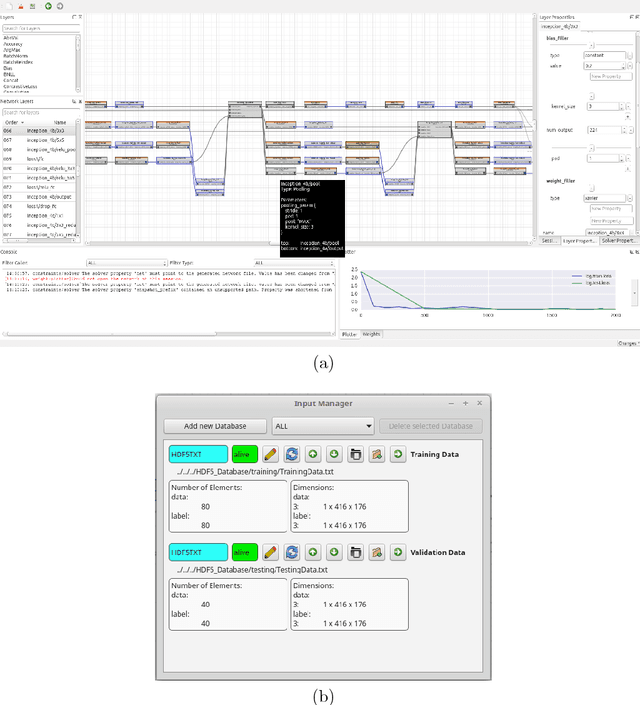
Abstract:In recent years, the importance of deep learning has significantly increased in pattern recognition, computer vision, and artificial intelligence research, as well as in industry. However, despite the existence of multiple deep learning frameworks, there is a lack of comprehensible and easy-to-use high-level tools for the design, training, and testing of deep neural networks (DNNs). In this paper, we introduce Barista, an open-source graphical high-level interface for the Caffe deep learning framework. While Caffe is one of the most popular frameworks for training DNNs, editing prototext files in order to specify the net architecture and hyper parameters can become a cumbersome and error-prone task. Instead, Barista offers a fully graphical user interface with a graph-based net topology editor and provides an end-to-end training facility for DNNs, which allows researchers to focus on solving their problems without having to write code, edit text files, or manually parse logged data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge