Udo Dannlowski
deepmriprep: Voxel-based Morphometry (VBM) Preprocessing via Deep Neural Networks
Aug 20, 2024



Abstract:Voxel-based Morphometry (VBM) has emerged as a powerful approach in neuroimaging research, utilized in over 7,000 studies since the year 2000. Using Magnetic Resonance Imaging (MRI) data, VBM assesses variations in the local density of brain tissue and examines its associations with biological and psychometric variables. Here, we present deepmriprep, a neural network-based pipeline that performs all necessary preprocessing steps for VBM analysis of T1-weighted MR images using deep neural networks. Utilizing the Graphics Processing Unit (GPU), deepmriprep is 37 times faster than CAT12, the leading VBM preprocessing toolbox. The proposed method matches CAT12 in accuracy for tissue segmentation and image registration across more than 100 datasets and shows strong correlations in VBM results. Tissue segmentation maps from deepmriprep have over 95% agreement with ground truth maps, and its non-linear registration, using supervised SYMNet, predicts smooth deformation fields comparable to CAT12. The high processing speed of deepmriprep enables rapid preprocessing of extensive datasets and thereby fosters the application of VBM analysis to large-scale neuroimaging studies and opens the door to real-time applications. Finally, deepmripreps straightforward, modular design enables researchers to easily understand, reuse, and advance the underlying methods, fostering further advancements in neuroimaging research. deepmriprep can be conveniently installed as a Python package and is publicly accessible at https://github.com/wwu-mmll/deepmriprep.
GateNet: A novel Neural Network Architecture for Automated Flow Cytometry Gating
Dec 12, 2023



Abstract:Flow cytometry is widely used to identify cell populations in patient-derived fluids such as peripheral blood (PB) or cerebrospinal fluid (CSF). While ubiquitous in research and clinical practice, flow cytometry requires gating, i.e. cell type identification which requires labor-intensive and error-prone manual adjustments. To facilitate this process, we designed GateNet, the first neural network architecture enabling full end-to-end automated gating without the need to correct for batch effects. We train GateNet with over 8,000,000 events based on N=127 PB and CSF samples which were manually labeled independently by four experts. We show that for novel, unseen samples, GateNet achieves human-level performance (F1 score ranging from 0.910 to 0.997). In addition we apply GateNet to a publicly available dataset confirming generalization with an F1 score of 0.936. As our implementation utilizes graphics processing units (GPU), gating only needs 15 microseconds per event. Importantly, we also show that GateNet only requires ~10 samples to reach human-level performance, rendering it widely applicable in all domains of flow cytometry.
DenseNet and Support Vector Machine classifications of major depressive disorder using vertex-wise cortical features
Nov 18, 2023



Abstract:Major depressive disorder (MDD) is a complex psychiatric disorder that affects the lives of hundreds of millions of individuals around the globe. Even today, researchers debate if morphological alterations in the brain are linked to MDD, likely due to the heterogeneity of this disorder. The application of deep learning tools to neuroimaging data, capable of capturing complex non-linear patterns, has the potential to provide diagnostic and predictive biomarkers for MDD. However, previous attempts to demarcate MDD patients and healthy controls (HC) based on segmented cortical features via linear machine learning approaches have reported low accuracies. In this study, we used globally representative data from the ENIGMA-MDD working group containing an extensive sample of people with MDD (N=2,772) and HC (N=4,240), which allows a comprehensive analysis with generalizable results. Based on the hypothesis that integration of vertex-wise cortical features can improve classification performance, we evaluated the classification of a DenseNet and a Support Vector Machine (SVM), with the expectation that the former would outperform the latter. As we analyzed a multi-site sample, we additionally applied the ComBat harmonization tool to remove potential nuisance effects of site. We found that both classifiers exhibited close to chance performance (balanced accuracy DenseNet: 51%; SVM: 53%), when estimated on unseen sites. Slightly higher classification performance (balanced accuracy DenseNet: 58%; SVM: 55%) was found when the cross-validation folds contained subjects from all sites, indicating site effect. In conclusion, the integration of vertex-wise morphometric features and the use of the non-linear classifier did not lead to the differentiability between MDD and HC. Our results support the notion that MDD classification on this combination of features and classifiers is unfeasible.
Deepbet: Fast brain extraction of T1-weighted MRI using Convolutional Neural Networks
Aug 14, 2023



Abstract:Brain extraction in magnetic resonance imaging (MRI) data is an important segmentation step in many neuroimaging preprocessing pipelines. Image segmentation is one of the research fields in which deep learning had the biggest impact in recent years enabling high precision segmentation with minimal compute. Consequently, traditional brain extraction methods are now being replaced by deep learning-based methods. Here, we used a unique dataset comprising 568 T1-weighted (T1w) MR images from 191 different studies in combination with cutting edge deep learning methods to build a fast, high-precision brain extraction tool called deepbet. deepbet uses LinkNet, a modern UNet architecture, in a two stage prediction process. This increases its segmentation performance, setting a novel state-of-the-art performance during cross-validation with a median Dice score (DSC) of 99.0% on unseen datasets, outperforming current state of the art models (DSC = 97.8% and DSC = 97.9%). While current methods are more sensitive to outliers, resulting in Dice scores as low as 76.5%, deepbet manages to achieve a Dice score of > 96.9% for all samples. Finally, our model accelerates brain extraction by a factor of ~10 compared to current methods, enabling the processing of one image in ~2 seconds on low level hardware.
From Group-Differences to Single-Subject Probability: Conformal Prediction-based Uncertainty Estimation for Brain-Age Modeling
Feb 10, 2023
Abstract:The brain-age gap is one of the most investigated risk markers for brain changes across disorders. While the field is progressing towards large-scale models, recently incorporating uncertainty estimates, no model to date provides the single-subject risk assessment capability essential for clinical application. In order to enable the clinical use of brain-age as a biomarker, we here combine uncertainty-aware deep Neural Networks with conformal prediction theory. This approach provides statistical guarantees with respect to single-subject uncertainty estimates and allows for the calculation of an individual's probability for accelerated brain-aging. Building on this, we show empirically in a sample of N=16,794 participants that 1. a lower or comparable error as state-of-the-art, large-scale brain-age models, 2. the statistical guarantees regarding single-subject uncertainty estimation indeed hold for every participant, and 3. that the higher individual probabilities of accelerated brain-aging derived from our model are associated with Alzheimer's Disease, Bipolar Disorder and Major Depressive Disorder.
An Uncertainty-Aware, Shareable and Transparent Neural Network Architecture for Brain-Age Modeling
Jul 16, 2021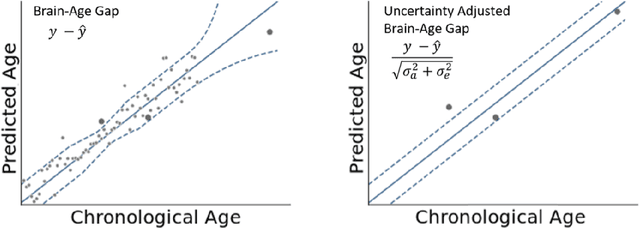
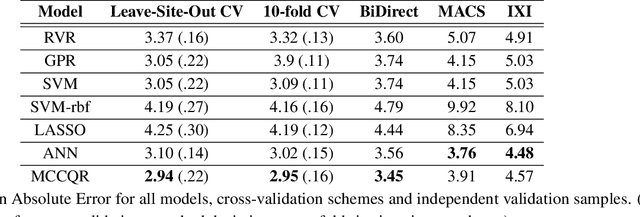
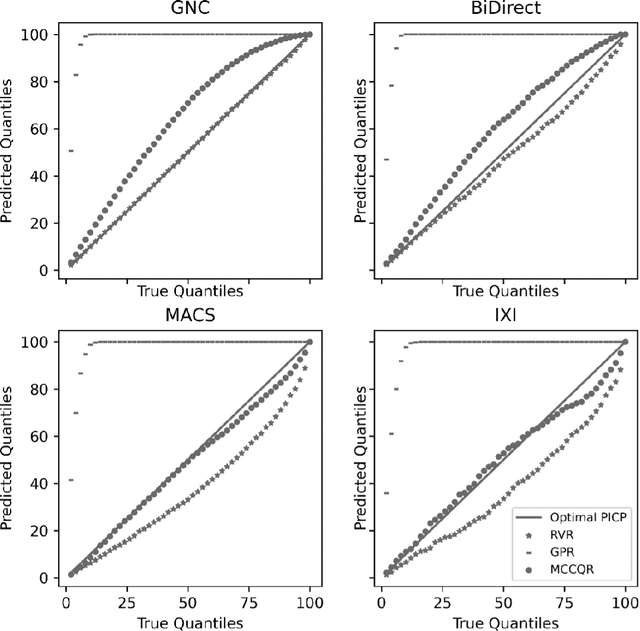
Abstract:The deviation between chronological age and age predicted from neuroimaging data has been identified as a sensitive risk-marker of cross-disorder brain changes, growing into a cornerstone of biological age-research. However, Machine Learning models underlying the field do not consider uncertainty, thereby confounding results with training data density and variability. Also, existing models are commonly based on homogeneous training sets, often not independently validated, and cannot be shared due to data protection issues. Here, we introduce an uncertainty-aware, shareable, and transparent Monte-Carlo Dropout Composite-Quantile-Regression (MCCQR) Neural Network trained on N=10,691 datasets from the German National Cohort. The MCCQR model provides robust, distribution-free uncertainty quantification in high-dimensional neuroimaging data, achieving lower error rates compared to existing models across ten recruitment centers and in three independent validation samples (N=4,004). In two examples, we demonstrate that it prevents spurious associations and increases power to detect accelerated brain-aging. We make the pre-trained model publicly available.
Predicting brain-age from raw T 1 -weighted Magnetic Resonance Imaging data using 3D Convolutional Neural Networks
Mar 22, 2021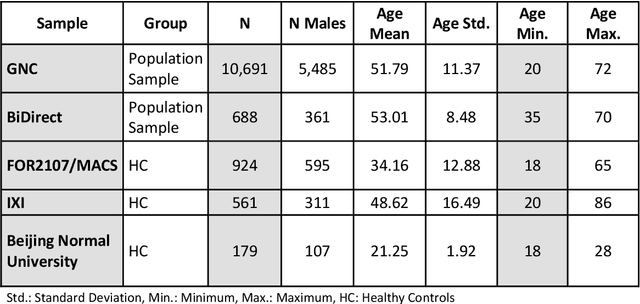
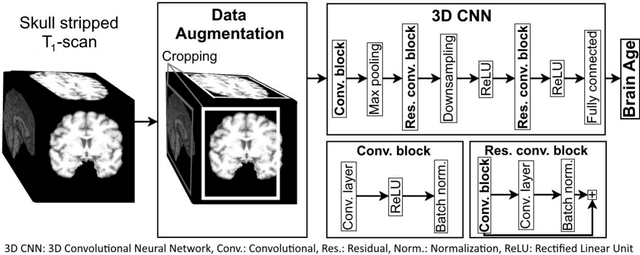
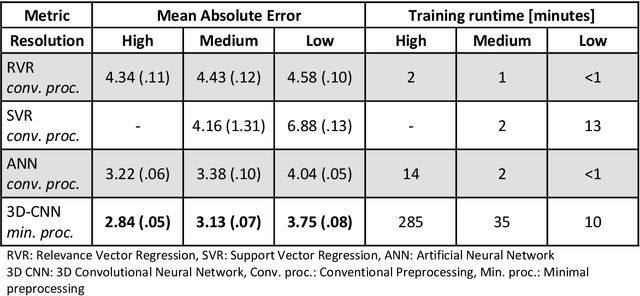
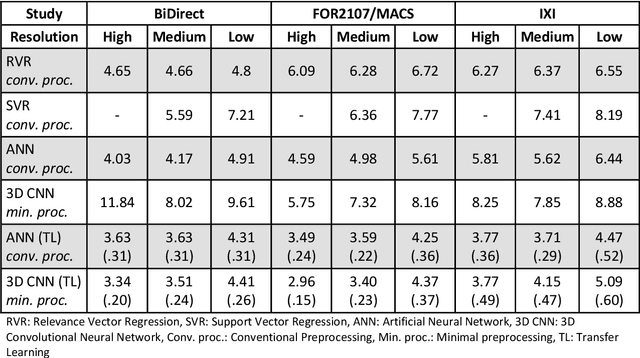
Abstract:Age prediction based on Magnetic Resonance Imaging (MRI) data of the brain is a biomarker to quantify the progress of brain diseases and aging. Current approaches rely on preparing the data with multiple preprocessing steps, such as registering voxels to a standardized brain atlas, which yields a significant computational overhead, hampers widespread usage and results in the predicted brain-age to be sensitive to preprocessing parameters. Here we describe a 3D Convolutional Neural Network (CNN) based on the ResNet architecture being trained on raw, non-registered T$_ 1$-weighted MRI data of N=10,691 samples from the German National Cohort and additionally applied and validated in N=2,173 samples from three independent studies using transfer learning. For comparison, state-of-the-art models using preprocessed neuroimaging data are trained and validated on the same samples. The 3D CNN using raw neuroimaging data predicts age with a mean average deviation of 2.84 years, outperforming the state-of-the-art brain-age models using preprocessed data. Since our approach is invariant to preprocessing software and parameter choices, it enables faster, more robust and more accurate brain-age modeling.
The PHOTON Wizard -- Towards Educational Machine Learning Code Generators
Feb 13, 2020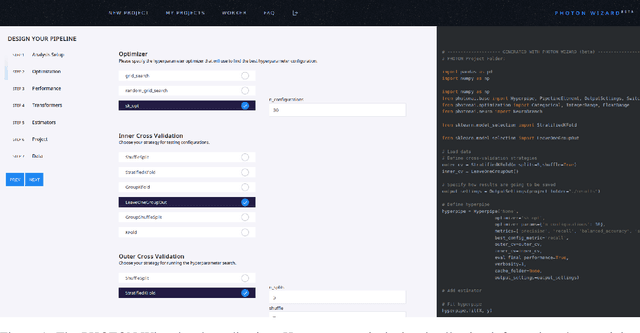
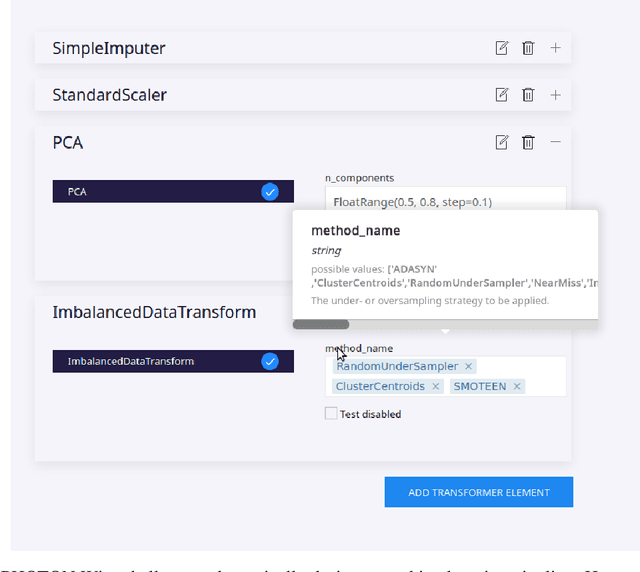
Abstract:Despite the tremendous efforts to democratize machine learning, especially in applied-science, the application is still often hampered by the lack of coding skills. As we consider programmatic understanding key to building effective and efficient machine learning solutions, we argue for a novel educational approach that builds upon the accessibility and acceptance of graphical user interfaces to convey programming skills to an applied-science target group. We outline a proof-of-concept, open-source web application, the PHOTON Wizard, which dynamically translates GUI interactions into valid source code for the Python machine learning framework PHOTON. Thereby, users possessing theoretical machine learning knowledge gain key insights into the model development workflow as well as an intuitive understanding of custom implementations. Specifically, the PHOTON Wizard integrates the concept of Educational Machine Learning Code Generators to teach users how to write code for designing, training, optimizing and evaluating custom machine learning pipelines.
PHOTON -- A Python API for Rapid Machine Learning Model Development
Feb 13, 2020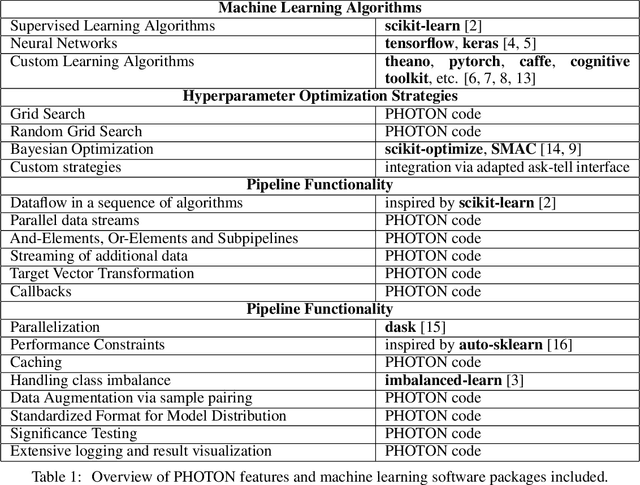
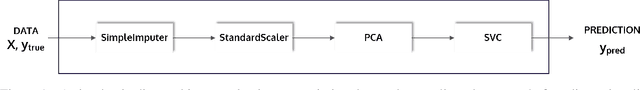
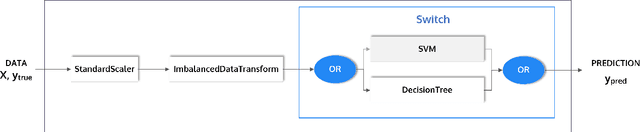
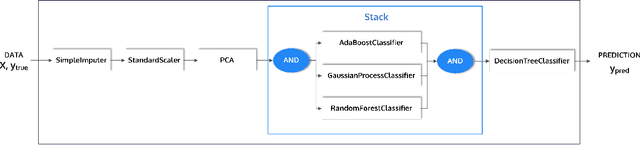
Abstract:This article describes the implementation and use of PHOTON, a high-level Python API designed to simplify and accelerate the process of machine learning model development. It enables designing both basic and advanced machine learning pipeline architectures and automatizes the repetitive training, optimization and evaluation workflow. PHOTON offers easy access to established machine learning toolboxes as well as the possibility to integrate custom algorithms and solutions for any part of the model construction and evaluation process. By adding a layer of abstraction incorporating current best practices it offers an easy-to-use, flexible approach to implementing fast, reproducible, and unbiased machine learning solutions.
Systematic Overestimation of Machine Learning Performance in Neuroimaging Studies of Depression
Dec 13, 2019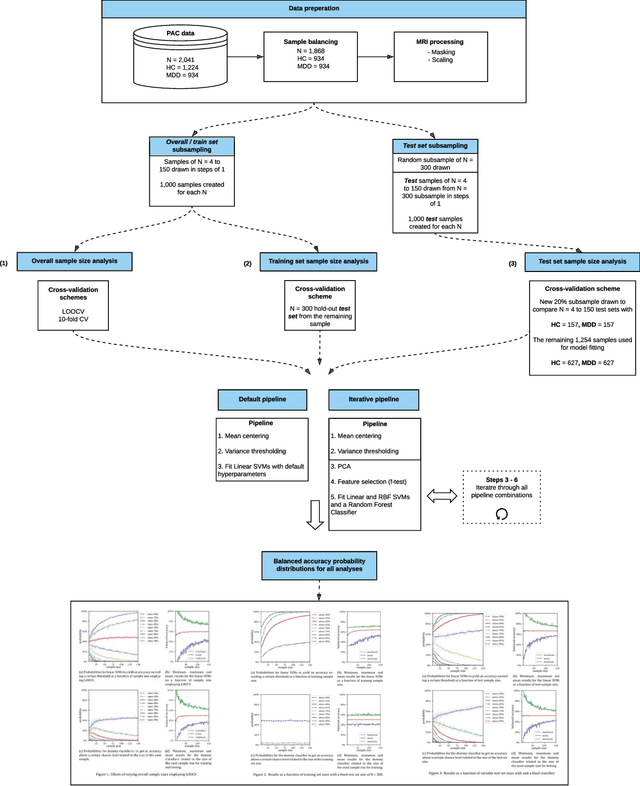
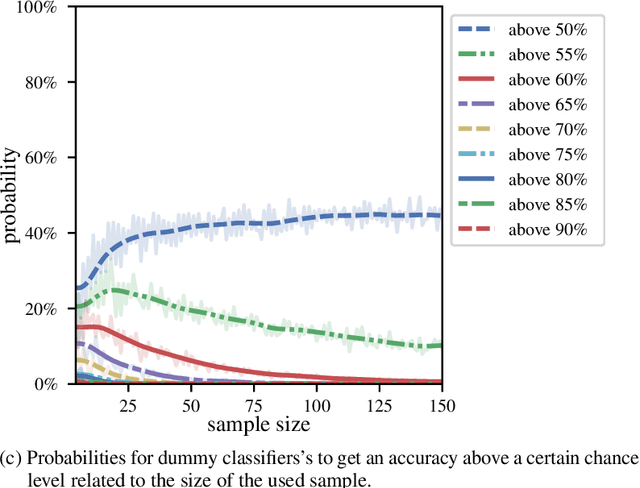
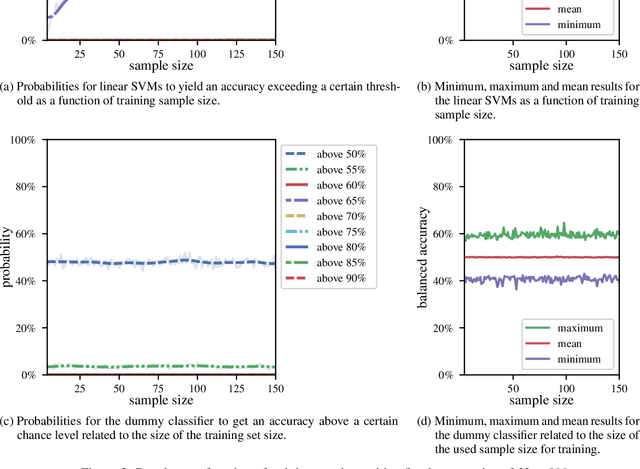
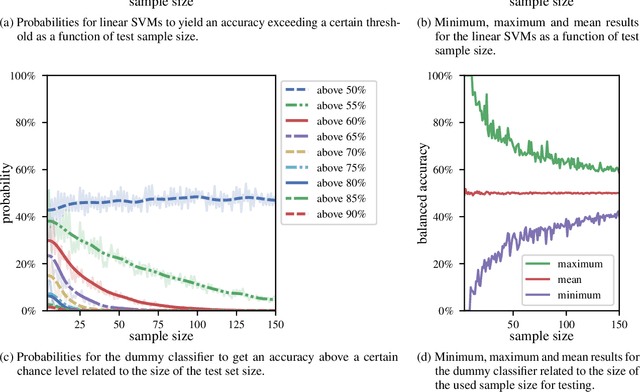
Abstract:We currently observe a disconcerting phenomenon in machine learning studies in psychiatry: While we would expect larger samples to yield better results due to the availability of more data, larger machine learning studies consistently show much weaker performance than the numerous small-scale studies. Here, we systematically investigated this effect focusing on one of the most heavily studied questions in the field, namely the classification of patients suffering from Major Depressive Disorder (MDD) and healthy controls. Drawing upon a balanced sample of $N = 1,868$ MDD patients and healthy controls from our recent international Predictive Analytics Competition (PAC), we first trained and tested a classification model on the full dataset which yielded an accuracy of 61%. Next, we mimicked the process by which researchers would draw samples of various sizes ($N=4$ to $N=150$) from the population and showed a strong risk of overestimation. Specifically, for small sample sizes ($N=20$), we observe accuracies of up to 95%. For medium sample sizes ($N=100$) accuracies up to 75% were found. Importantly, further investigation showed that sufficiently large test sets effectively protect against performance overestimation whereas larger datasets per se do not. While these results question the validity of a substantial part of the current literature, we outline the relatively low-cost remedy of larger test sets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge