Xiaofeng Liu
Structure-aware Unsupervised Tagged-to-Cine MRI Synthesis with Self Disentanglement
Feb 25, 2022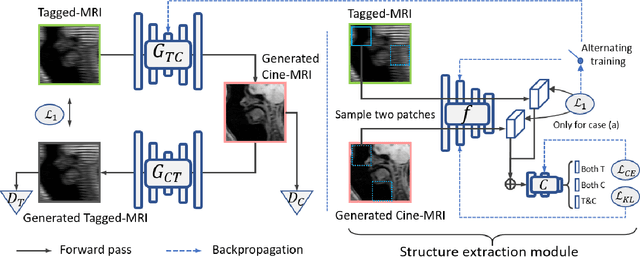
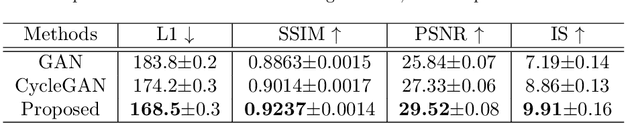
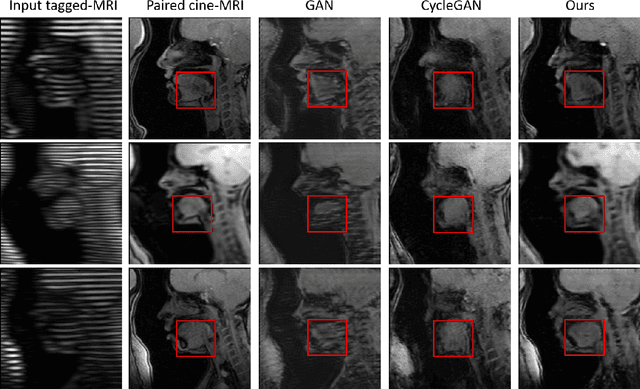
Abstract:Cycle reconstruction regularized adversarial training -- e.g., CycleGAN, DiscoGAN, and DualGAN -- has been widely used for image style transfer with unpaired training data. Several recent works, however, have shown that local distortions are frequent, and structural consistency cannot be guaranteed. Targeting this issue, prior works usually relied on additional segmentation or consistent feature extraction steps that are task-specific. To counter this, this work aims to learn a general add-on structural feature extractor, by explicitly enforcing the structural alignment between an input and its synthesized image. Specifically, we propose a novel input-output image patches self-training scheme to achieve a disentanglement of underlying anatomical structures and imaging modalities. The translator and structure encoder are updated, following an alternating training protocol. In addition, the information w.r.t. imaging modality can be eliminated with an asymmetric adversarial game. We train, validate, and test our network on 1,768, 416, and 1,560 unpaired subject-independent slices of tagged and cine magnetic resonance imaging from a total of twenty healthy subjects, respectively, demonstrating superior performance over competing methods.
A Multi-modal Fusion Framework Based on Multi-task Correlation Learning for Cancer Prognosis Prediction
Jan 22, 2022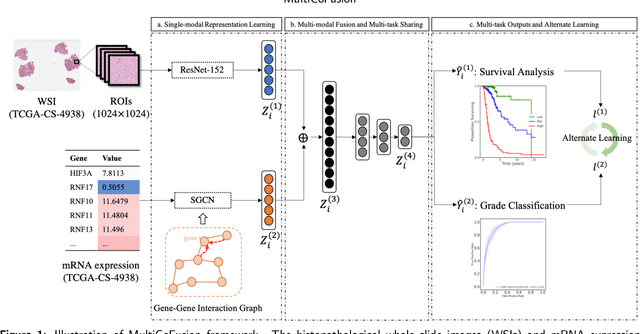
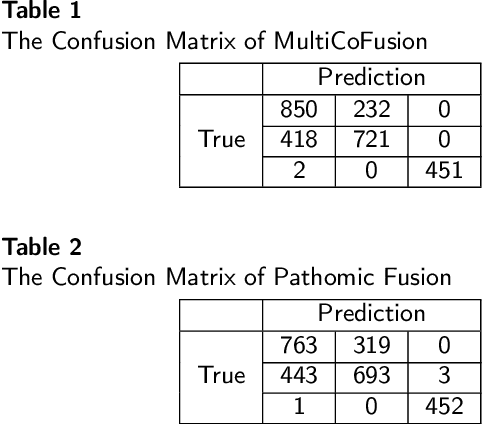
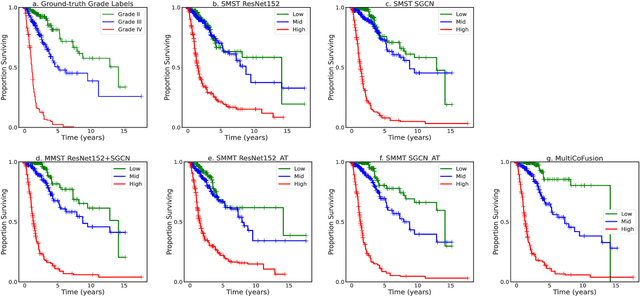
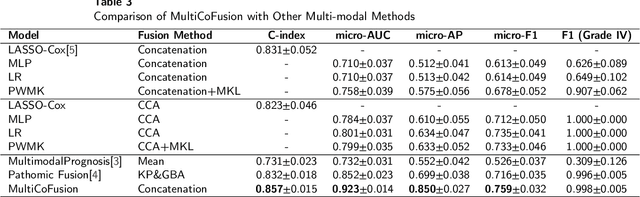
Abstract:Morphological attributes from histopathological images and molecular profiles from genomic data are important information to drive diagnosis, prognosis, and therapy of cancers. By integrating these heterogeneous but complementary data, many multi-modal methods are proposed to study the complex mechanisms of cancers, and most of them achieve comparable or better results from previous single-modal methods. However, these multi-modal methods are restricted to a single task (e.g., survival analysis or grade classification), and thus neglect the correlation between different tasks. In this study, we present a multi-modal fusion framework based on multi-task correlation learning (MultiCoFusion) for survival analysis and cancer grade classification, which combines the power of multiple modalities and multiple tasks. Specifically, a pre-trained ResNet-152 and a sparse graph convolutional network (SGCN) are used to learn the representations of histopathological images and mRNA expression data respectively. Then these representations are fused by a fully connected neural network (FCNN), which is also a multi-task shared network. Finally, the results of survival analysis and cancer grade classification output simultaneously. The framework is trained by an alternate scheme. We systematically evaluate our framework using glioma datasets from The Cancer Genome Atlas (TCGA). Results demonstrate that MultiCoFusion learns better representations than traditional feature extraction methods. With the help of multi-task alternating learning, even simple multi-modal concatenation can achieve better performance than other deep learning and traditional methods. Multi-task learning can improve the performance of multiple tasks not just one of them, and it is effective in both single-modal and multi-modal data.
Variational Inference for Quantifying Inter-observer Variability in Segmentation of Anatomical Structures
Jan 18, 2022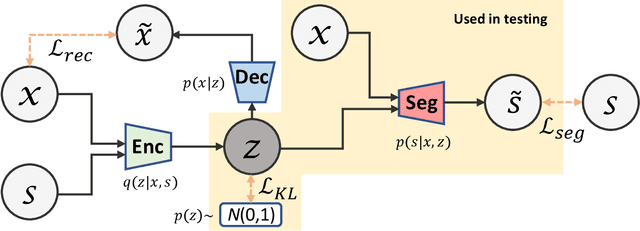

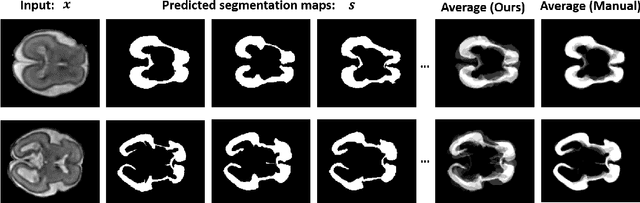

Abstract:Lesions or organ boundaries visible through medical imaging data are often ambiguous, thus resulting in significant variations in multi-reader delineations, i.e., the source of aleatoric uncertainty. In particular, quantifying the inter-observer variability of manual annotations with Magnetic Resonance (MR) Imaging data plays a crucial role in establishing a reference standard for various diagnosis and treatment tasks. Most segmentation methods, however, simply model a mapping from an image to its single segmentation map and do not take the disagreement of annotators into consideration. In order to account for inter-observer variability, without sacrificing accuracy, we propose a novel variational inference framework to model the distribution of plausible segmentation maps, given a specific MR image, which explicitly represents the multi-reader variability. Specifically, we resort to a latent vector to encode the multi-reader variability and counteract the inherent information loss in the imaging data. Then, we apply a variational autoencoder network and optimize its evidence lower bound (ELBO) to efficiently approximate the distribution of the segmentation map, given an MR image. Experimental results, carried out with the QUBIQ brain growth MRI segmentation datasets with seven annotators, demonstrate the effectiveness of our approach.
Self-semantic contour adaptation for cross modality brain tumor segmentation
Jan 13, 2022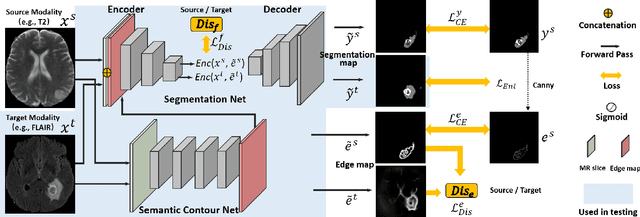
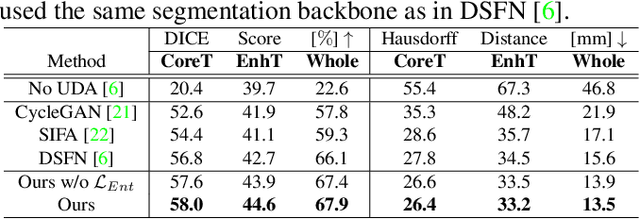
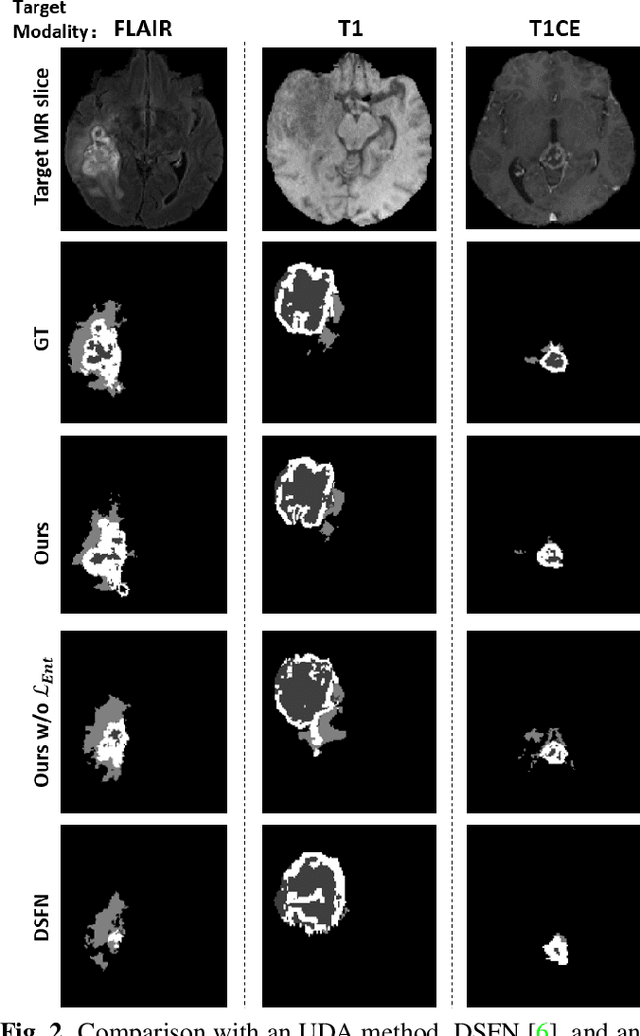
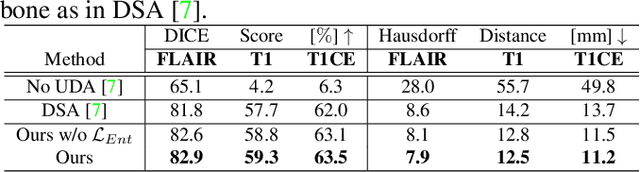
Abstract:Unsupervised domain adaptation (UDA) between two significantly disparate domains to learn high-level semantic alignment is a crucial yet challenging task.~To this end, in this work, we propose exploiting low-level edge information to facilitate the adaptation as a precursor task, which has a small cross-domain gap, compared with semantic segmentation.~The precise contour then provides spatial information to guide the semantic adaptation. More specifically, we propose a multi-task framework to learn a contouring adaptation network along with a semantic segmentation adaptation network, which takes both magnetic resonance imaging (MRI) slice and its initial edge map as input.~These two networks are jointly trained with source domain labels, and the feature and edge map level adversarial learning is carried out for cross-domain alignment. In addition, self-entropy minimization is incorporated to further enhance segmentation performance. We evaluated our framework on the BraTS2018 database for cross-modality segmentation of brain tumors, showing the validity and superiority of our approach, compared with competing methods.
Surrogate Model for Shallow Water Equations Solvers with Deep Learning
Dec 20, 2021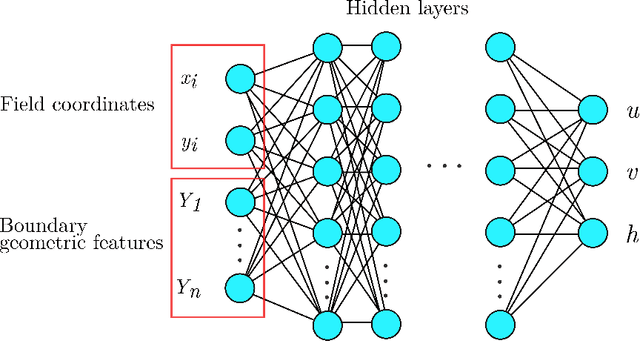
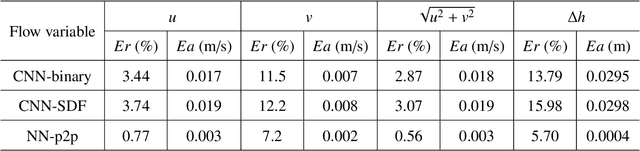
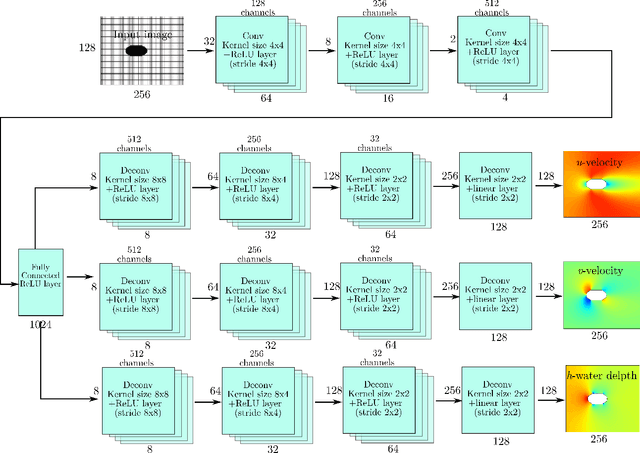
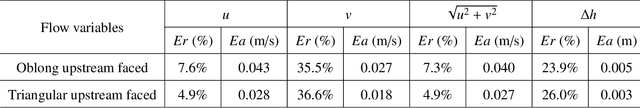
Abstract:Shallow water equations are the foundation of most models for flooding and river hydraulics analysis. These physics-based models are usually expensive and slow to run, thus not suitable for real-time prediction or parameter inversion. An attractive alternative is surrogate model. This work introduces an efficient, accurate, and flexible surrogate model, NN-p2p, based on deep learning and it can make point-to-point predictions on unstructured or irregular meshes. The new method was evaluated and compared against existing methods based on convolutional neural networks (CNNs), which can only make image-to-image predictions on structured or regular meshes. In NN-p2p, the input includes both spatial coordinates and boundary features that can describe the geometry of hydraulic structures, such as bridge piers. All surrogate models perform well in predicting flow around different types of piers in the training domain. However, only NN-p2p works well when spatial extrapolation is performed. The limitations of CNN-based methods are rooted in their raster-image nature which cannot capture boundary geometry and flow features exactly, which are of paramount importance to fluid dynamics. NN-p2p also has good performance in predicting flow around piers unseen by the neural network. The NN-p2p model also respects conservation laws more strictly. The application of the proposed surrogate model was demonstrated by calculating the drag coefficient $C_D$ for piers and a new linear relationship between $C_D$ and the logarithmic transformation of pier's length/width ratio was discovered.
Recursively Conditional Gaussian for Ordinal Unsupervised Domain Adaptation
Aug 17, 2021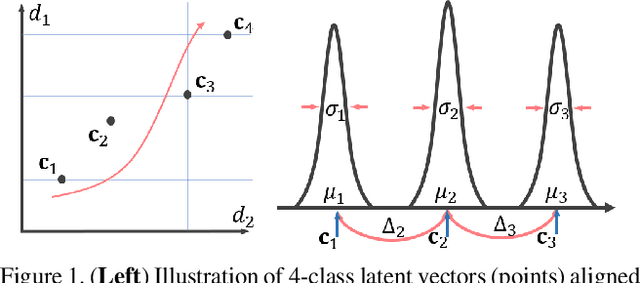
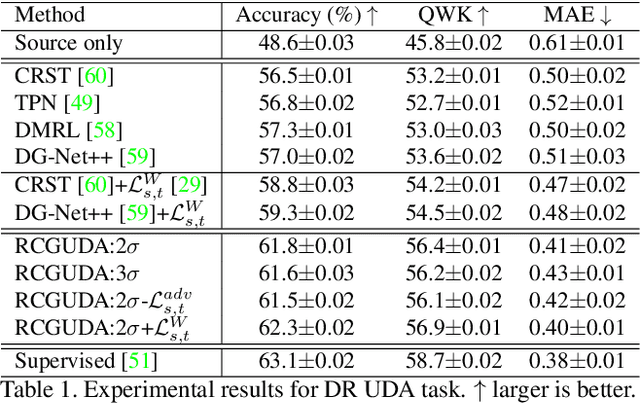
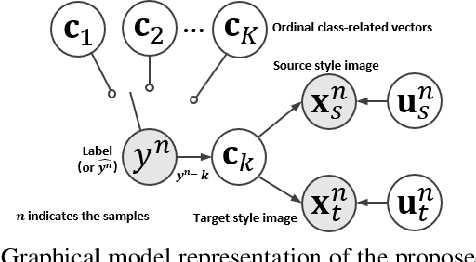
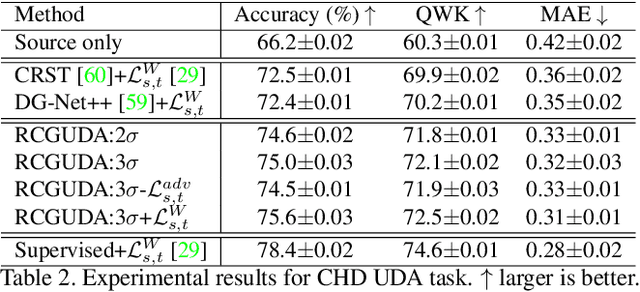
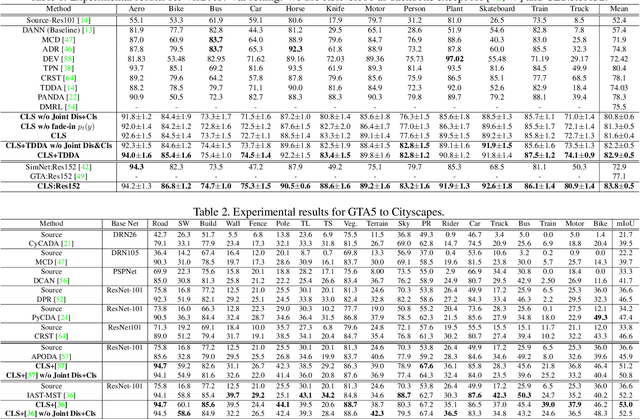
Abstract:The unsupervised domain adaptation (UDA) has been widely adopted to alleviate the data scalability issue, while the existing works usually focus on classifying independently discrete labels. However, in many tasks (e.g., medical diagnosis), the labels are discrete and successively distributed. The UDA for ordinal classification requires inducing non-trivial ordinal distribution prior to the latent space. Target for this, the partially ordered set (poset) is defined for constraining the latent vector. Instead of the typically i.i.d. Gaussian latent prior, in this work, a recursively conditional Gaussian (RCG) set is adapted for ordered constraint modeling, which admits a tractable joint distribution prior. Furthermore, we are able to control the density of content vector that violates the poset constraints by a simple "three-sigma rule". We explicitly disentangle the cross-domain images into a shared ordinal prior induced ordinal content space and two separate source/target ordinal-unrelated spaces, and the self-training is worked on the shared space exclusively for ordinal-aware domain alignment. Extensive experiments on UDA medical diagnoses and facial age estimation demonstrate its effectiveness.
Adversarial Unsupervised Domain Adaptation with Conditional and Label Shift: Infer, Align and Iterate
Aug 02, 2021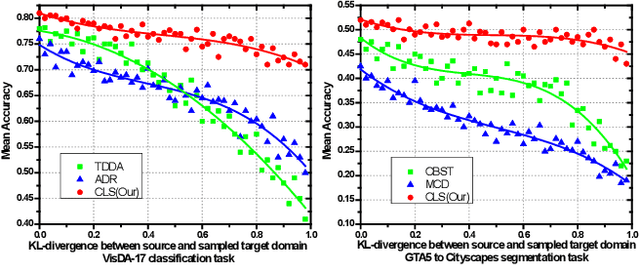
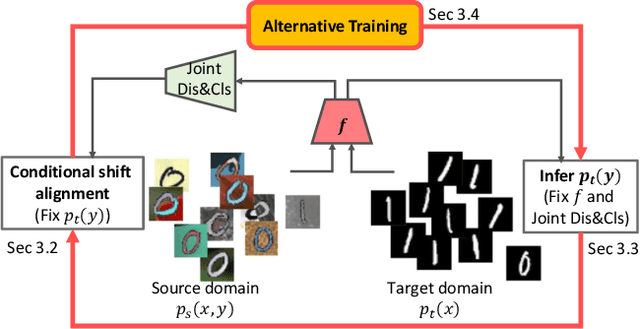
Abstract:In this work, we propose an adversarial unsupervised domain adaptation (UDA) approach with the inherent conditional and label shifts, in which we aim to align the distributions w.r.t. both $p(x|y)$ and $p(y)$. Since the label is inaccessible in the target domain, the conventional adversarial UDA assumes $p(y)$ is invariant across domains, and relies on aligning $p(x)$ as an alternative to the $p(x|y)$ alignment. To address this, we provide a thorough theoretical and empirical analysis of the conventional adversarial UDA methods under both conditional and label shifts, and propose a novel and practical alternative optimization scheme for adversarial UDA. Specifically, we infer the marginal $p(y)$ and align $p(x|y)$ iteratively in the training, and precisely align the posterior $p(y|x)$ in testing. Our experimental results demonstrate its effectiveness on both classification and segmentation UDA, and partial UDA.
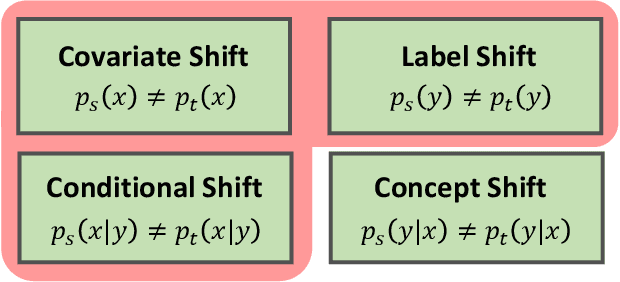
Domain Generalization under Conditional and Label Shifts via Variational Bayesian Inference
Jul 22, 2021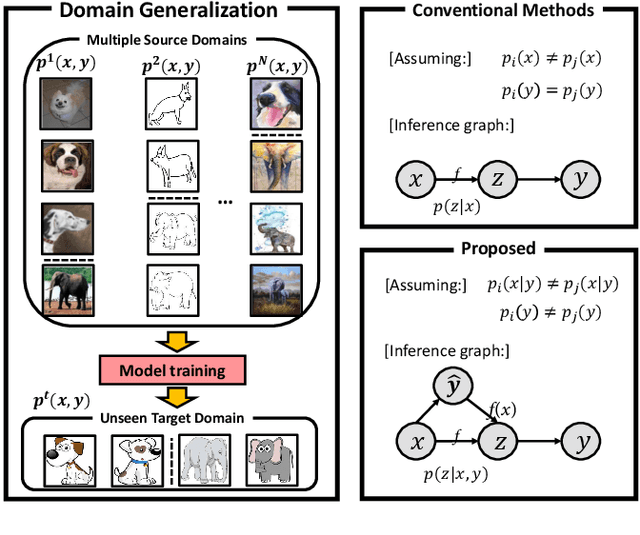
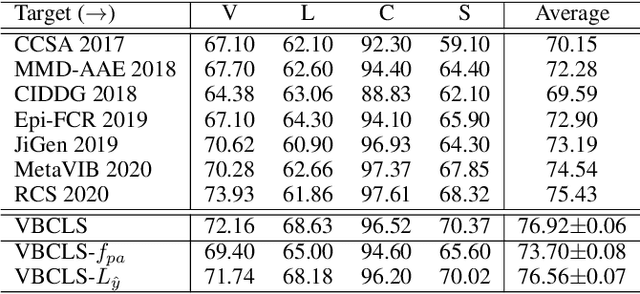
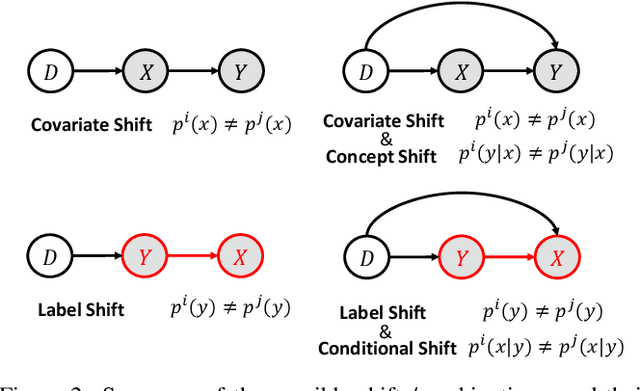
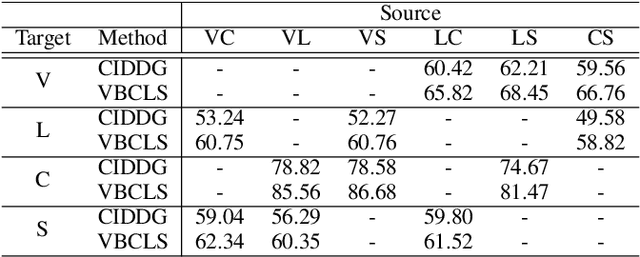
Abstract:In this work, we propose a domain generalization (DG) approach to learn on several labeled source domains and transfer knowledge to a target domain that is inaccessible in training. Considering the inherent conditional and label shifts, we would expect the alignment of $p(x|y)$ and $p(y)$. However, the widely used domain invariant feature learning (IFL) methods relies on aligning the marginal concept shift w.r.t. $p(x)$, which rests on an unrealistic assumption that $p(y)$ is invariant across domains. We thereby propose a novel variational Bayesian inference framework to enforce the conditional distribution alignment w.r.t. $p(x|y)$ via the prior distribution matching in a latent space, which also takes the marginal label shift w.r.t. $p(y)$ into consideration with the posterior alignment. Extensive experiments on various benchmarks demonstrate that our framework is robust to the label shift and the cross-domain accuracy is significantly improved, thereby achieving superior performance over the conventional IFL counterparts.
Segmentation of Cardiac Structures via Successive Subspace Learning with Saab Transform from Cine MRI
Jul 22, 2021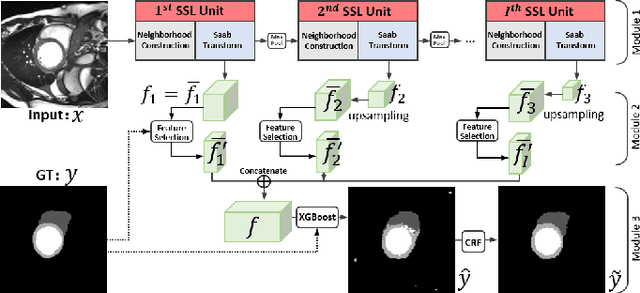
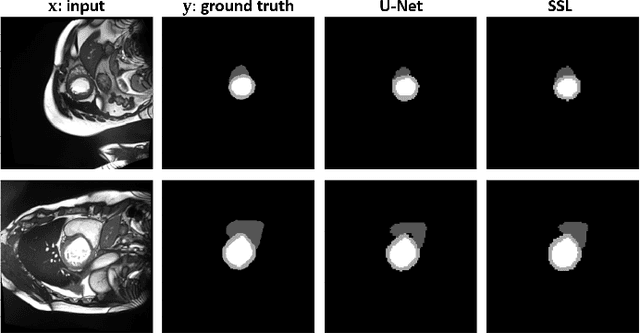
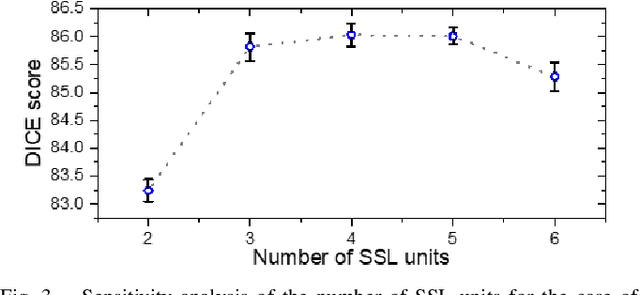
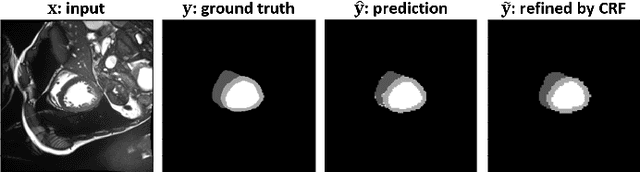
Abstract:Assessment of cardiovascular disease (CVD) with cine magnetic resonance imaging (MRI) has been used to non-invasively evaluate detailed cardiac structure and function. Accurate segmentation of cardiac structures from cine MRI is a crucial step for early diagnosis and prognosis of CVD, and has been greatly improved with convolutional neural networks (CNN). There, however, are a number of limitations identified in CNN models, such as limited interpretability and high complexity, thus limiting their use in clinical practice. In this work, to address the limitations, we propose a lightweight and interpretable machine learning model, successive subspace learning with the subspace approximation with adjusted bias (Saab) transform, for accurate and efficient segmentation from cine MRI. Specifically, our segmentation framework is comprised of the following steps: (1) sequential expansion of near-to-far neighborhood at different resolutions; (2) channel-wise subspace approximation using the Saab transform for unsupervised dimension reduction; (3) class-wise entropy guided feature selection for supervised dimension reduction; (4) concatenation of features and pixel-wise classification with gradient boost; and (5) conditional random field for post-processing. Experimental results on the ACDC 2017 segmentation database, showed that our framework performed better than state-of-the-art U-Net models with 200$\times$ fewer parameters in delineating the left ventricle, right ventricle, and myocardium, thus showing its potential to be used in clinical practice.
Deep 3D-CNN for Depression Diagnosis with Facial Video Recording of Self-Rating Depression Scale Questionnaire
Jul 22, 2021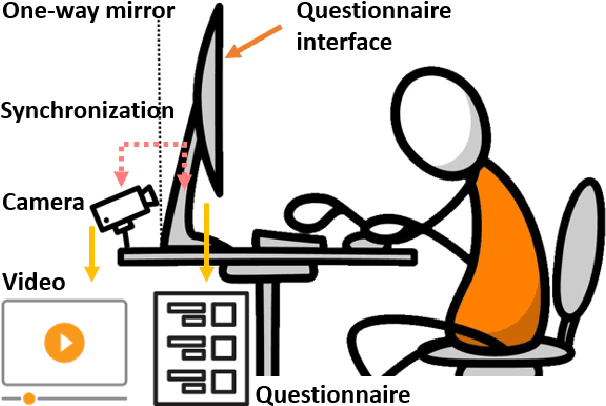
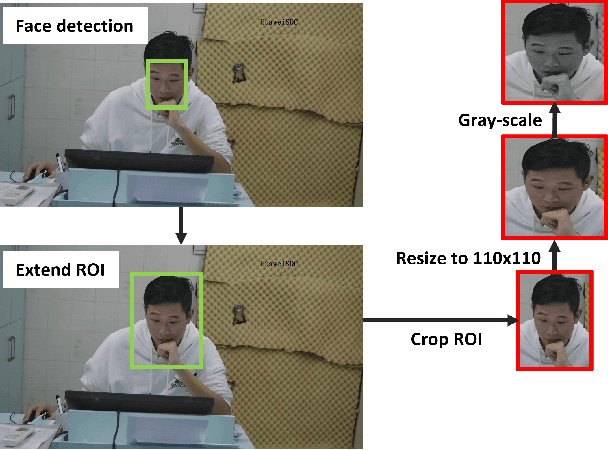
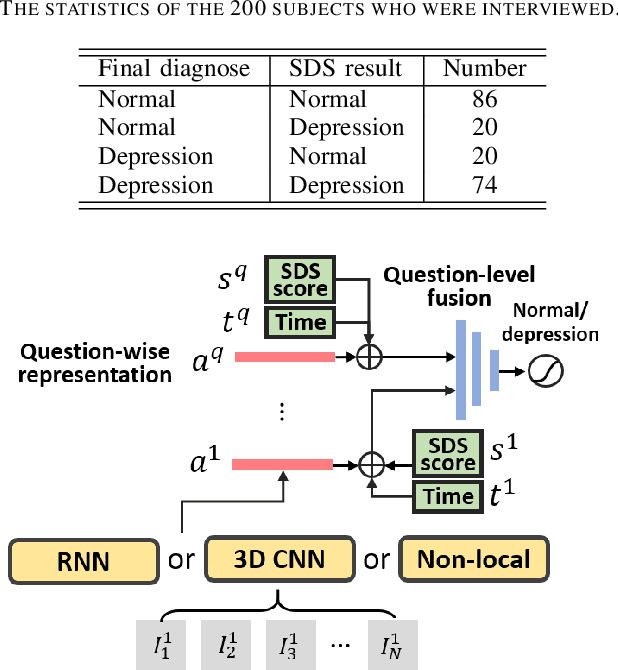
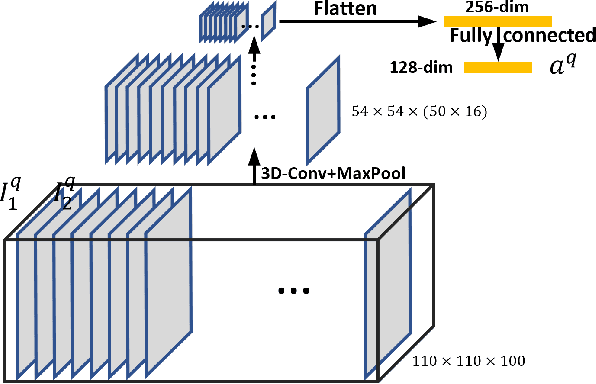
Abstract:The Self-Rating Depression Scale (SDS) questionnaire is commonly utilized for effective depression preliminary screening. The uncontrolled self-administered measure, on the other hand, maybe readily influenced by insouciant or dishonest responses, yielding different findings from the clinician-administered diagnostic. Facial expression (FE) and behaviors are important in clinician-administered assessments, but they are underappreciated in self-administered evaluations. We use a new dataset of 200 participants to demonstrate the validity of self-rating questionnaires and their accompanying question-by-question video recordings in this study. We offer an end-to-end system to handle the face video recording that is conditioned on the questionnaire answers and the responding time to automatically interpret sadness from the SDS assessment and the associated video. We modified a 3D-CNN for temporal feature extraction and compared various state-of-the-art temporal modeling techniques. The superior performance of our system shows the validity of combining facial video recording with the SDS score for more accurate self-diagnose.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge