V. K. Cody Bumgardner
A Framework for Cross-Domain Generalization in Coronary Artery Calcium Scoring Across Gated and Non-Gated Computed Tomography
Feb 25, 2026Abstract:Coronary artery calcium (CAC) scoring is a key predictor of cardiovascular risk, but it relies on ECG-gated CT scans, restricting its use to specialized cardiac imaging settings. We introduce an automated framework for CAC detection and lesion-specific Agatston scoring that operates across both gated and non-gated CT scans. At its core is CARD-ViT, a self-supervised Vision Transformer trained exclusively on gated CT data using DINO. Without any non-gated training data, our framework achieves 0.707 accuracy and a Cohen's kappa of 0.528 on the Stanford non-gated dataset, matching models trained directly on non-gated scans. On gated test sets, the framework achieves 0.910 accuracy with Cohen's kappa scores of 0.871 and 0.874 across independent datasets, demonstrating robust risk stratification. These results demonstrate the feasibility of cross-domain CAC scoring from gated to non-gated domains, supporting scalable cardiovascular screening in routine chest imaging without additional scans or annotations.
Magnification-Aware Distillation (MAD): A Self-Supervised Framework for Unified Representation Learning in Gigapixel Whole-Slide Images
Dec 16, 2025Abstract:Whole-slide images (WSIs) contain tissue information distributed across multiple magnification levels, yet most self-supervised methods treat these scales as independent views. This separation prevents models from learning representations that remain stable when resolution changes, a key requirement for practical neuropathology workflows. This study introduces Magnification-Aware Distillation (MAD), a self-supervised strategy that links low-magnification context with spatially aligned high-magnification detail, enabling the model to learn how coarse tissue structure relates to fine cellular patterns. The resulting foundation model, MAD-NP, is trained entirely through this cross-scale correspondence without annotations. A linear classifier trained only on 10x embeddings maintains 96.7% of its performance when applied to unseen 40x tiles, demonstrating strong resolution-invariant representation learning. Segmentation outputs remain consistent across magnifications, preserving anatomical boundaries and minimizing noise. These results highlight the feasibility of scalable, magnification-robust WSI analysis using a unified embedding space
Toward an AI Reasoning-Enabled System for Patient-Clinical Trial Matching
Dec 08, 2025Abstract:Screening patients for clinical trial eligibility remains a manual, time-consuming, and resource-intensive process. We present a secure, scalable proof-of-concept system for Artificial Intelligence (AI)-augmented patient-trial matching that addresses key implementation challenges: integrating heterogeneous electronic health record (EHR) data, facilitating expert review, and maintaining rigorous security standards. Leveraging open-source, reasoning-enabled large language models (LLMs), the system moves beyond binary classification to generate structured eligibility assessments with interpretable reasoning chains that support human-in-the-loop review. This decision support tool represents eligibility as a dynamic state rather than a fixed determination, identifying matches when available and offering actionable recommendations that could render a patient eligible in the future. The system aims to reduce coordinator burden, intelligently broaden the set of trials considered for each patient and guarantee comprehensive auditability of all AI-generated outputs.
Bridging the Clinical Expertise Gap: Development of a Web-Based Platform for Accessible Time Series Forecasting and Analysis
Dec 08, 2025



Abstract:Time series forecasting has applications across domains and industries, especially in healthcare, but the technical expertise required to analyze data, build models, and interpret results can be a barrier to using these techniques. This article presents a web platform that makes the process of analyzing and plotting data, training forecasting models, and interpreting and viewing results accessible to researchers and clinicians. Users can upload data and generate plots to showcase their variables and the relationships between them. The platform supports multiple forecasting models and training techniques which are highly customizable according to the user's needs. Additionally, recommendations and explanations can be generated from a large language model that can help the user choose appropriate parameters for their data and understand the results for each model. The goal is to integrate this platform into learning health systems for continuous data collection and inference from clinical pipelines.
Forecasting Opioid Incidents for Rapid Actionable Data for Opioid Response in Kentucky
Oct 21, 2024Abstract:We present efforts in the fields of machine learning and time series forecasting to accurately predict counts of future opioid overdose incidents recorded by Emergency Medical Services (EMS) in the state of Kentucky. Forecasts are useful to state government agencies to properly prepare and distribute resources related to opioid overdoses effectively. Our approach uses county and district level aggregations of EMS opioid overdose encounters and forecasts future counts for each month. A variety of additional covariates were tested to determine their impact on the model's performance. Models with different levels of complexity were evaluated to optimize training time and accuracy. Our results show that when special precautions are taken to address data sparsity, useful predictions can be generated with limited error by utilizing yearly trends and covariance with additional data sources.
Institutional Platform for Secure Self-Service Large Language Model Exploration
Feb 01, 2024



Abstract:This paper introduces a user-friendly platform developed by the University of Kentucky Center for Applied AI, designed to make large, customized language models (LLMs) more accessible. By capitalizing on recent advancements in multi-LoRA inference, the system efficiently accommodates custom adapters for a diverse range of users and projects. The paper outlines the system's architecture and key features, encompassing dataset curation, model training, secure inference, and text-based feature extraction. We illustrate the establishment of a tenant-aware computational network using agent-based methods, securely utilizing islands of isolated resources as a unified system. The platform strives to deliver secure LLM services, emphasizing process and data isolation, end-to-end encryption, and role-based resource authentication. This contribution aligns with the overarching goal of enabling simplified access to cutting-edge AI models and technology in support of scientific discovery.
Multi-Modal Machine Learning Framework for Automated Seizure Detection in Laboratory Rats
Feb 01, 2024Abstract:A multi-modal machine learning system uses multiple unique data sources and types to improve its performance. This article proposes a system that combines results from several types of models, all of which are trained on different data signals. As an example to illustrate the efficacy of the system, an experiment is described in which multiple types of data are collected from rats suffering from seizures. This data includes electrocorticography readings, piezoelectric motion sensor data, and video recordings. Separate models are trained on each type of data, with the goal of classifying each time frame as either containing a seizure or not. After each model has generated its classification predictions, these results are combined. While each data signal works adequately on its own for prediction purposes, the significant imbalance in class labels leads to increased numbers of false positives, which can be filtered and removed by utilizing all data sources. This paper will demonstrate that, after postprocessing and combination techniques, classification accuracy is improved with this multi-modal system when compared to the performance of each individual data source.
CLASSify: A Web-Based Tool for Machine Learning
Oct 05, 2023Abstract:Machine learning classification problems are widespread in bioinformatics, but the technical knowledge required to perform model training, optimization, and inference can prevent researchers from utilizing this technology. This article presents an automated tool for machine learning classification problems to simplify the process of training models and producing results while providing informative visualizations and insights into the data. This tool supports both binary and multiclass classification problems, and it provides access to a variety of models and methods. Synthetic data can be generated within the interface to fill missing values, balance class labels, or generate entirely new datasets. It also provides support for feature evaluation and generates explainability scores to indicate which features influence the output the most. We present CLASSify, an open-source tool for simplifying the user experience of solving classification problems without the need for knowledge of machine learning.
Local Large Language Models for Complex Structured Medical Tasks
Aug 03, 2023Abstract:This paper introduces an approach that combines the language reasoning capabilities of large language models (LLMs) with the benefits of local training to tackle complex, domain-specific tasks. Specifically, the authors demonstrate their approach by extracting structured condition codes from pathology reports. The proposed approach utilizes local LLMs, which can be fine-tuned to respond to specific generative instructions and provide structured outputs. The authors collected a dataset of over 150k uncurated surgical pathology reports, containing gross descriptions, final diagnoses, and condition codes. They trained different model architectures, including LLaMA, BERT and LongFormer and evaluated their performance. The results show that the LLaMA-based models significantly outperform BERT-style models across all evaluated metrics, even with extremely reduced precision. The LLaMA models performed especially well with large datasets, demonstrating their ability to handle complex, multi-label tasks. Overall, this work presents an effective approach for utilizing LLMs to perform domain-specific tasks using accessible hardware, with potential applications in the medical domain, where complex data extraction and classification are required.
Semantic Enrichment of Streaming Healthcare Data
Dec 01, 2019
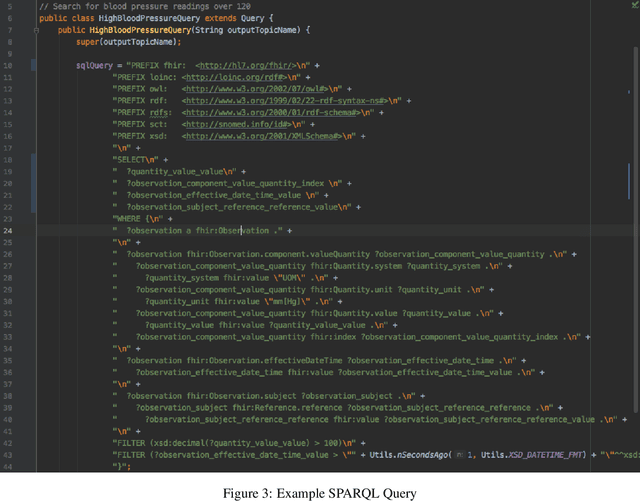
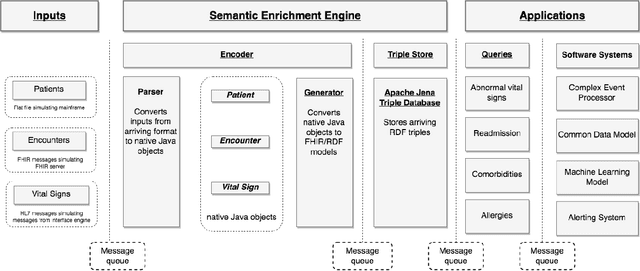
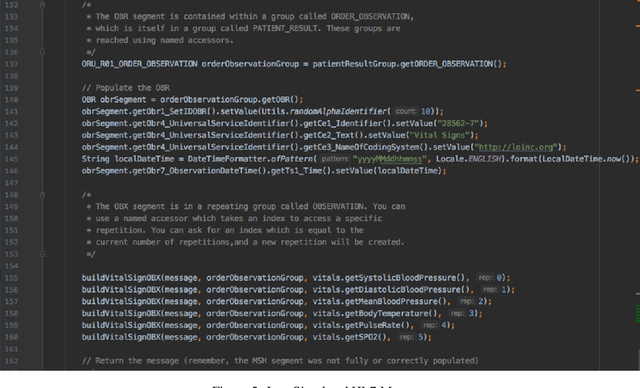
Abstract:In the past decade, the healthcare industry has made significant advances in the digitization of patient information. However, a lack of interoperability among healthcare systems still imposes a high cost to patients, hospitals, and insurers. Currently, most systems pass messages using idiosyncratic messaging standards that require specialized knowledge to interpret. This increases the cost of systems integration and often puts more advanced uses of data out of reach. In this project, we demonstrate how two open standards, FHIR and RDF, can be combined both to integrate data from disparate sources in real-time and make that data queryable and susceptible to automated inference. To validate the effectiveness of the semantic engine, we perform simulations of real-time data feeds and demonstrate how they can be combined and used by client-side applications with no knowledge of the underlying sources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge