David A Gutman
Magnification-Aware Distillation (MAD): A Self-Supervised Framework for Unified Representation Learning in Gigapixel Whole-Slide Images
Dec 16, 2025Abstract:Whole-slide images (WSIs) contain tissue information distributed across multiple magnification levels, yet most self-supervised methods treat these scales as independent views. This separation prevents models from learning representations that remain stable when resolution changes, a key requirement for practical neuropathology workflows. This study introduces Magnification-Aware Distillation (MAD), a self-supervised strategy that links low-magnification context with spatially aligned high-magnification detail, enabling the model to learn how coarse tissue structure relates to fine cellular patterns. The resulting foundation model, MAD-NP, is trained entirely through this cross-scale correspondence without annotations. A linear classifier trained only on 10x embeddings maintains 96.7% of its performance when applied to unseen 40x tiles, demonstrating strong resolution-invariant representation learning. Segmentation outputs remain consistent across magnifications, preserving anatomical boundaries and minimizing noise. These results highlight the feasibility of scalable, magnification-robust WSI analysis using a unified embedding space
HistomicsML2.0: Fast interactive machine learning for whole slide imaging data
Jan 30, 2020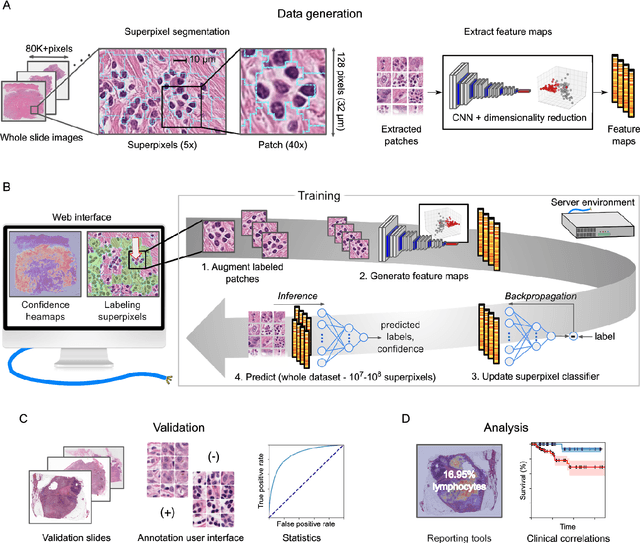
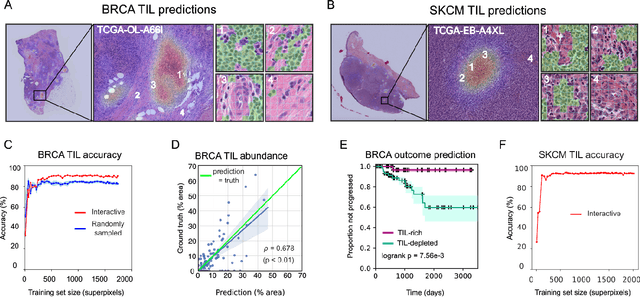
Abstract:Extracting quantitative phenotypic information from whole-slide images presents significant challenges for investigators who are not experienced in developing image analysis algorithms. We present new software that enables rapid learn-by-example training of machine learning classifiers for detection of histologic patterns in whole-slide imaging datasets. HistomicsML2.0 uses convolutional networks to be readily adaptable to a variety of applications, provides a web-based user interface, and is available as a software container to simplify deployment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge