Tilo Burghardt
Training dynamics in Physics-Informed Neural Networks with feature mapping
Feb 10, 2024Abstract:Physics-Informed Neural Networks (PINNs) have emerged as an iconic machine learning approach for solving Partial Differential Equations (PDEs). Although its variants have achieved significant progress, the empirical success of utilising feature mapping from the wider Implicit Neural Representations studies has been substantially neglected. We investigate the training dynamics of PINNs with a feature mapping layer via the limiting Conjugate Kernel and Neural Tangent Kernel, which sheds light on the convergence and generalisation of the model. We also show the inadequacy of commonly used Fourier-based feature mapping in some scenarios and propose the conditional positive definite Radial Basis Function as a better alternative. The empirical results reveal the efficacy of our method in diverse forward and inverse problem sets. This simple technique can be easily implemented in coordinate input networks and benefits the broad PINNs research.
PanAf20K: A Large Video Dataset for Wild Ape Detection and Behaviour Recognition
Jan 31, 2024



Abstract:We present the PanAf20K dataset, the largest and most diverse open-access annotated video dataset of great apes in their natural environment. It comprises more than 7 million frames across ~20,000 camera trap videos of chimpanzees and gorillas collected at 14 field sites in tropical Africa as part of the Pan African Programme: The Cultured Chimpanzee. The footage is accompanied by a rich set of annotations and benchmarks making it suitable for training and testing a variety of challenging and ecologically important computer vision tasks including ape detection and behaviour recognition. Furthering AI analysis of camera trap information is critical given the International Union for Conservation of Nature now lists all species in the great ape family as either Endangered or Critically Endangered. We hope the dataset can form a solid basis for engagement of the AI community to improve performance, efficiency, and result interpretation in order to support assessments of great ape presence, abundance, distribution, and behaviour and thereby aid conservation efforts.
Diving with Penguins: Detecting Penguins and their Prey in Animal-borne Underwater Videos via Deep Learning
Aug 14, 2023



Abstract:African penguins (Spheniscus demersus) are an endangered species. Little is known regarding their underwater hunting strategies and associated predation success rates, yet this is essential for guiding conservation. Modern bio-logging technology has the potential to provide valuable insights, but manually analysing large amounts of data from animal-borne video recorders (AVRs) is time-consuming. In this paper, we publish an animal-borne underwater video dataset of penguins and introduce a ready-to-deploy deep learning system capable of robustly detecting penguins (mAP50@98.0%) and also instances of fish (mAP50@73.3%). We note that the detectors benefit explicitly from air-bubble learning to improve accuracy. Extending this detector towards a dual-stream behaviour recognition network, we also provide the first results for identifying predation behaviour in penguin underwater videos. Whilst results are promising, further work is required for useful applicability of predation behaviour detection in field scenarios. In summary, we provide a highly reliable underwater penguin detector, a fish detector, and a valuable first attempt towards an automated visual detection of complex behaviours in a marine predator. We publish the networks, the DivingWithPenguins video dataset, annotations, splits, and weights for full reproducibility and immediate usability by practitioners.
Deep Visual-Genetic Biometrics for Taxonomic Classification of Rare Species
May 20, 2023



Abstract:Visual as well as genetic biometrics are routinely employed to identify species and individuals in biological applications. However, no attempts have been made in this domain to computationally enhance visual classification of rare classes with little image data via genetics. In this paper, we thus propose aligned visual-genetic inference spaces with the aim to implicitly encode cross-domain associations for improved performance. We demonstrate for the first time that such alignment can be achieved via deep embedding models and that the approach is directly applicable to boosting long-tailed recognition (LTR) particularly for rare species. We experimentally demonstrate the efficacy of the concept via application to microscopic imagery of 30k+ planktic foraminifer shells across 32 species when used together with independent genetic data samples. Most importantly for practitioners, we show that visual-genetic alignment can significantly benefit visual-only recognition of the rarest species. Technically, we pre-train a visual ResNet50 deep learning model using triplet loss formulations to create an initial embedding space. We re-structure this space based on genetic anchors embedded via a Sequence Graph Transform (SGT) and linked to visual data by cross-domain cosine alignment. We show that an LTR approach improves the state-of-the-art across all benchmarks and that adding our visual-genetic alignment improves per-class and particularly rare tail class benchmarks significantly further. We conclude that visual-genetic alignment can be a highly effective tool for complementing visual biological data containing rare classes. The concept proposed may serve as an important future tool for integrating genetics and imageomics towards a more complete scientific representation of taxonomic spaces and life itself. Code, weights, and data splits are published for full reproducibility.
Use Your Head: Improving Long-Tail Video Recognition
Apr 03, 2023



Abstract:This paper presents an investigation into long-tail video recognition. We demonstrate that, unlike naturally-collected video datasets and existing long-tail image benchmarks, current video benchmarks fall short on multiple long-tailed properties. Most critically, they lack few-shot classes in their tails. In response, we propose new video benchmarks that better assess long-tail recognition, by sampling subsets from two datasets: SSv2 and VideoLT. We then propose a method, Long-Tail Mixed Reconstruction, which reduces overfitting to instances from few-shot classes by reconstructing them as weighted combinations of samples from head classes. LMR then employs label mixing to learn robust decision boundaries. It achieves state-of-the-art average class accuracy on EPIC-KITCHENS and the proposed SSv2-LT and VideoLT-LT. Benchmarks and code at: tobyperrett.github.io/lmr
Video-SwinUNet: Spatio-temporal Deep Learning Framework for VFSS Instance Segmentation
Feb 22, 2023Abstract:This paper presents a deep learning framework for medical video segmentation. Convolution neural network (CNN) and transformer-based methods have achieved great milestones in medical image segmentation tasks due to their incredible semantic feature encoding and global information comprehension abilities. However, most existing approaches ignore a salient aspect of medical video data - the temporal dimension. Our proposed framework explicitly extracts features from neighbouring frames across the temporal dimension and incorporates them with a temporal feature blender, which then tokenises the high-level spatio-temporal feature to form a strong global feature encoded via a Swin Transformer. The final segmentation results are produced via a UNet-like encoder-decoder architecture. Our model outperforms other approaches by a significant margin and improves the segmentation benchmarks on the VFSS2022 dataset, achieving a dice coefficient of 0.8986 and 0.8186 for the two datasets tested. Our studies also show the efficacy of the temporal feature blending scheme and cross-dataset transferability of learned capabilities. Code and models are fully available at https://github.com/SimonZeng7108/Video-SwinUNet.
Triple-stream Deep Metric Learning of Great Ape Behavioural Actions
Jan 06, 2023



Abstract:We propose the first metric learning system for the recognition of great ape behavioural actions. Our proposed triple stream embedding architecture works on camera trap videos taken directly in the wild and demonstrates that the utilisation of an explicit DensePose-C chimpanzee body part segmentation stream effectively complements traditional RGB appearance and optical flow streams. We evaluate system variants with different feature fusion techniques and long-tail recognition approaches. Results and ablations show performance improvements of ~12% in top-1 accuracy over previous results achieved on the PanAf-500 dataset containing 180,000 manually annotated frames across nine behavioural actions. Furthermore, we provide a qualitative analysis of our findings and augment the metric learning system with long-tail recognition techniques showing that average per class accuracy -- critical in the domain -- can be improved by ~23% compared to the literature on that dataset. Finally, since our embedding spaces are constructed as metric, we provide first data-driven visualisations of the great ape behavioural action spaces revealing emerging geometry and topology. We hope that the work sparks further interest in this vital application area of computer vision for the benefit of endangered great apes.
Video-TransUNet: Temporally Blended Vision Transformer for CT VFSS Instance Segmentation
Aug 22, 2022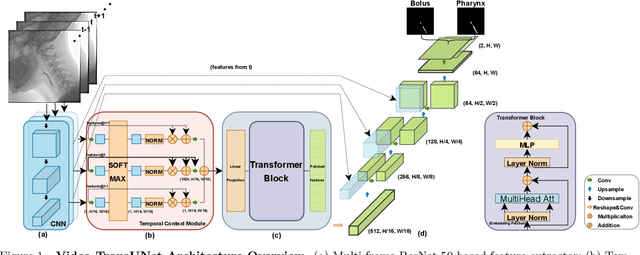
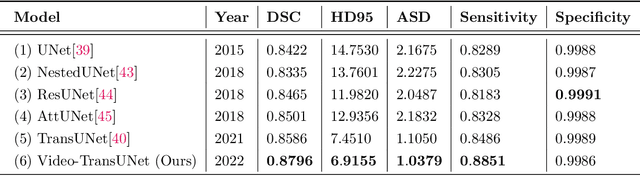

Abstract:We propose Video-TransUNet, a deep architecture for instance segmentation in medical CT videos constructed by integrating temporal feature blending into the TransUNet deep learning framework. In particular, our approach amalgamates strong frame representation via a ResNet CNN backbone, multi-frame feature blending via a Temporal Context Module (TCM), non-local attention via a Vision Transformer, and reconstructive capabilities for multiple targets via a UNet-based convolutional-deconvolutional architecture with multiple heads. We show that this new network design can significantly outperform other state-of-the-art systems when tested on the segmentation of bolus and pharynx/larynx in Videofluoroscopic Swallowing Study (VFSS) CT sequences. On our VFSS2022 dataset it achieves a dice coefficient of 0.8796 and an average surface distance of 1.0379 pixels. Note that tracking the pharyngeal bolus accurately is a particularly important application in clinical practice since it constitutes the primary method for diagnostics of swallowing impairment. Our findings suggest that the proposed model can indeed enhance the TransUNet architecture via exploiting temporal information and improving segmentation performance by a significant margin. We publish key source code, network weights, and ground truth annotations for simplified performance reproduction.
Inertial Hallucinations -- When Wearable Inertial Devices Start Seeing Things
Jul 14, 2022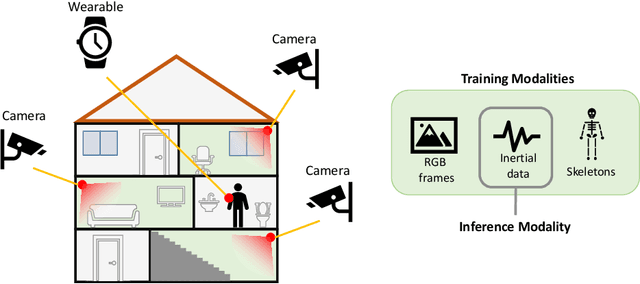
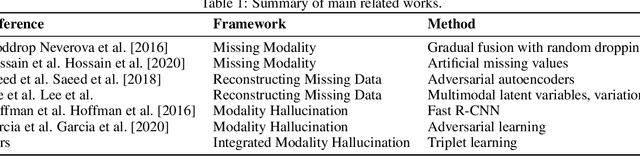
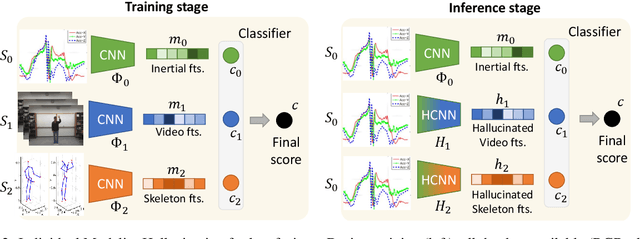
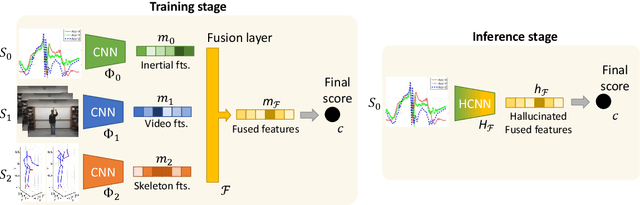
Abstract:We propose a novel approach to multimodal sensor fusion for Ambient Assisted Living (AAL) which takes advantage of learning using privileged information (LUPI). We address two major shortcomings of standard multimodal approaches, limited area coverage and reduced reliability. Our new framework fuses the concept of modality hallucination with triplet learning to train a model with different modalities to handle missing sensors at inference time. We evaluate the proposed model on inertial data from a wearable accelerometer device, using RGB videos and skeletons as privileged modalities, and show an improvement of accuracy of an average 6.6% on the UTD-MHAD dataset and an average 5.5% on the Berkeley MHAD dataset, reaching a new state-of-the-art for inertial-only classification accuracy on these datasets. We validate our framework through several ablation studies.
Towards Individual Grevy's Zebra Identification via Deep 3D Fitting and Metric Learning
Jun 07, 2022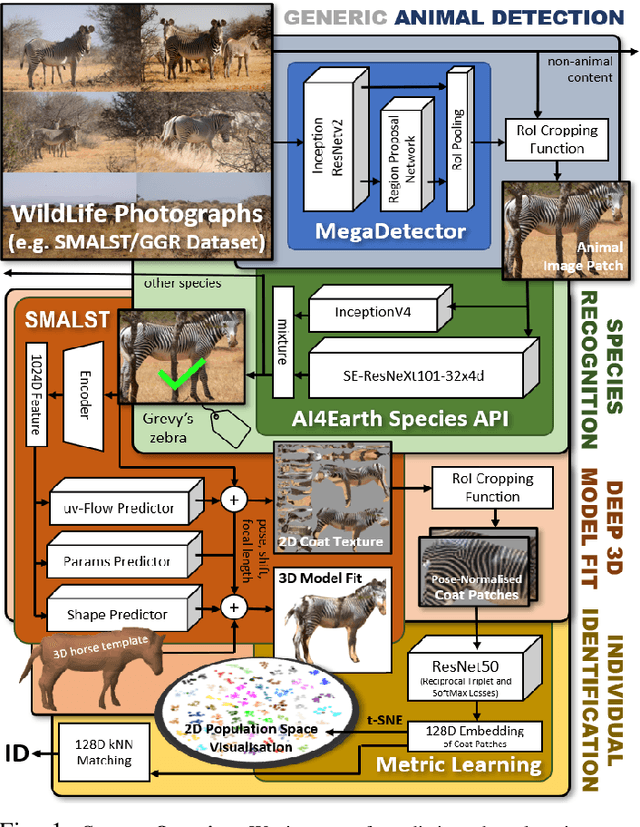
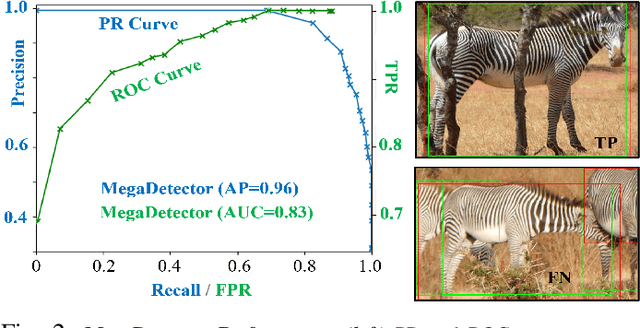
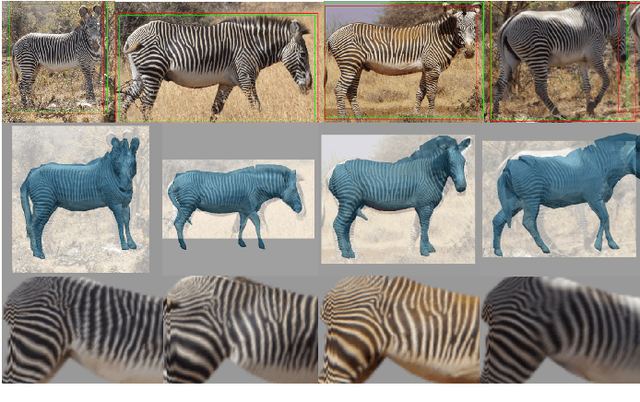
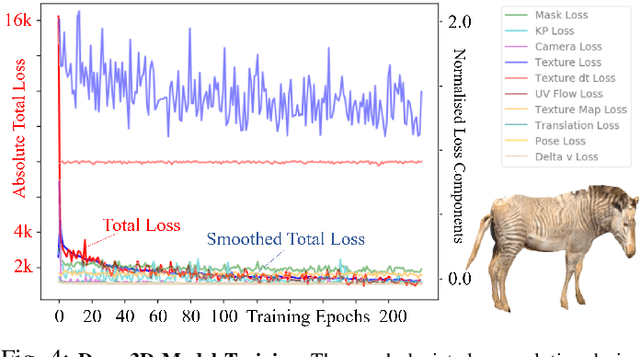
Abstract:This paper combines deep learning techniques for species detection, 3D model fitting, and metric learning in one pipeline to perform individual animal identification from photographs by exploiting unique coat patterns. This is the first work to attempt this and, compared to traditional 2D bounding box or segmentation based CNN identification pipelines, the approach provides effective and explicit view-point normalisation and allows for a straight forward visualisation of the learned biometric population space. Note that due to the use of metric learning the pipeline is also readily applicable to open set and zero shot re-identification scenarios. We apply the proposed approach to individual Grevy's zebra (Equus grevyi) identification and show in a small study on the SMALST dataset that the use of 3D model fitting can indeed benefit performance. In particular, back-projected textures from 3D fitted models improve identification accuracy from 48.0% to 56.8% compared to 2D bounding box approaches for the dataset. Whilst the study is far too small accurately to estimate the full performance potential achievable in larger-scale real-world application settings and in comparisons against polished tools, our work lays the conceptual and practical foundations for a next step in animal biometrics towards deep metric learning driven, fully 3D-aware animal identification in open population settings. We publish network weights and relevant facilitating source code with this paper for full reproducibility and as inspiration for further research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge