Daniel I. Rubenstein
kabr-tools: Automated Framework for Multi-Species Behavioral Monitoring
Oct 02, 2025Abstract:A comprehensive understanding of animal behavior ecology depends on scalable approaches to quantify and interpret complex, multidimensional behavioral patterns. Traditional field observations are often limited in scope, time-consuming, and labor-intensive, hindering the assessment of behavioral responses across landscapes. To address this, we present kabr-tools (Kenyan Animal Behavior Recognition Tools), an open-source package for automated multi-species behavioral monitoring. This framework integrates drone-based video with machine learning systems to extract behavioral, social, and spatial metrics from wildlife footage. Our pipeline leverages object detection, tracking, and behavioral classification systems to generate key metrics, including time budgets, behavioral transitions, social interactions, habitat associations, and group composition dynamics. Compared to ground-based methods, drone-based observations significantly improved behavioral granularity, reducing visibility loss by 15% and capturing more transitions with higher accuracy and continuity. We validate kabr-tools through three case studies, analyzing 969 behavioral sequences, surpassing the capacity of traditional methods for data capture and annotation. We found that, like Plains zebras, vigilance in Grevy's zebras decreases with herd size, but, unlike Plains zebras, habitat has a negligible impact. Plains and Grevy's zebras exhibit strong behavioral inertia, with rare transitions to alert behaviors and observed spatial segregation between Grevy's zebras, Plains zebras, and giraffes in mixed-species herds. By enabling automated behavioral monitoring at scale, kabr-tools offers a powerful tool for ecosystem-wide studies, advancing conservation, biodiversity research, and ecological monitoring.
HotSpotter - Patterned Species Instance Recognition
Aug 25, 2025Abstract:We present HotSpotter, a fast, accurate algorithm for identifying individual animals against a labeled database. It is not species specific and has been applied to Grevy's and plains zebras, giraffes, leopards, and lionfish. We describe two approaches, both based on extracting and matching keypoints or "hotspots". The first tests each new query image sequentially against each database image, generating a score for each database image in isolation, and ranking the results. The second, building on recent techniques for instance recognition, matches the query image against the database using a fast nearest neighbor search. It uses a competitive scoring mechanism derived from the Local Naive Bayes Nearest Neighbor algorithm recently proposed for category recognition. We demonstrate results on databases of more than 1000 images, producing more accurate matches than published methods and matching each query image in just a few seconds.
* Original matlab code: https://github.com/Erotemic/hotspotter-matlab-2013, Python port: https://github.com/Erotemic/hotspotter
Discovering Novel Biological Traits From Images Using Phylogeny-Guided Neural Networks
Jun 05, 2023



Abstract:Discovering evolutionary traits that are heritable across species on the tree of life (also referred to as a phylogenetic tree) is of great interest to biologists to understand how organisms diversify and evolve. However, the measurement of traits is often a subjective and labor-intensive process, making trait discovery a highly label-scarce problem. We present a novel approach for discovering evolutionary traits directly from images without relying on trait labels. Our proposed approach, Phylo-NN, encodes the image of an organism into a sequence of quantized feature vectors -- or codes -- where different segments of the sequence capture evolutionary signals at varying ancestry levels in the phylogeny. We demonstrate the effectiveness of our approach in producing biologically meaningful results in a number of downstream tasks including species image generation and species-to-species image translation, using fish species as a target example.
Towards Individual Grevy's Zebra Identification via Deep 3D Fitting and Metric Learning
Jun 07, 2022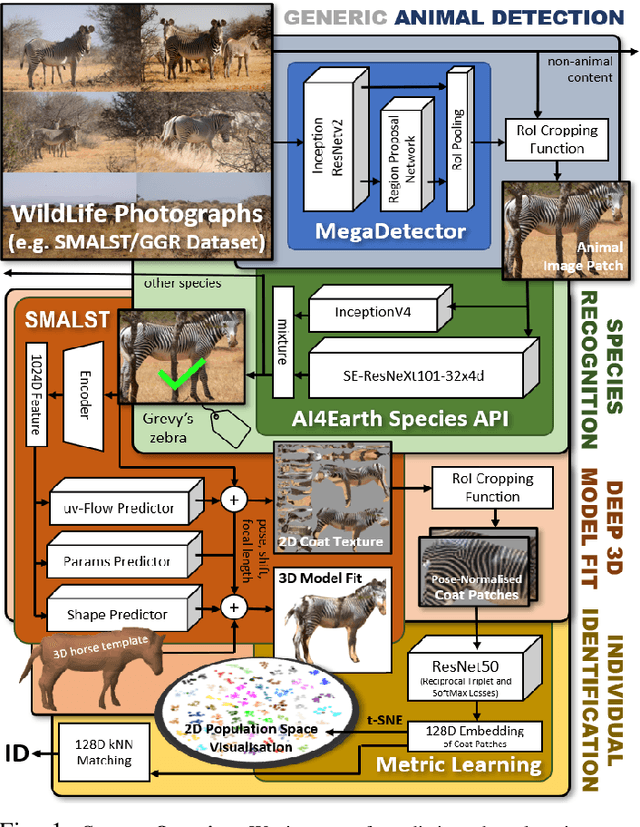
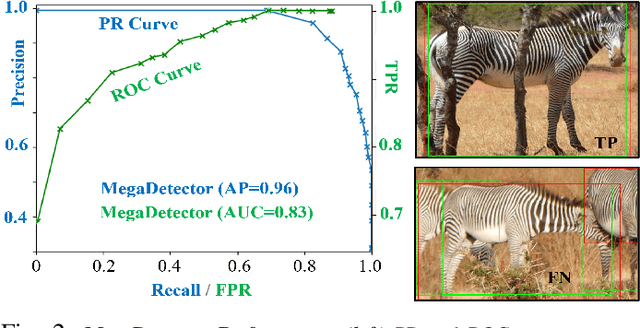
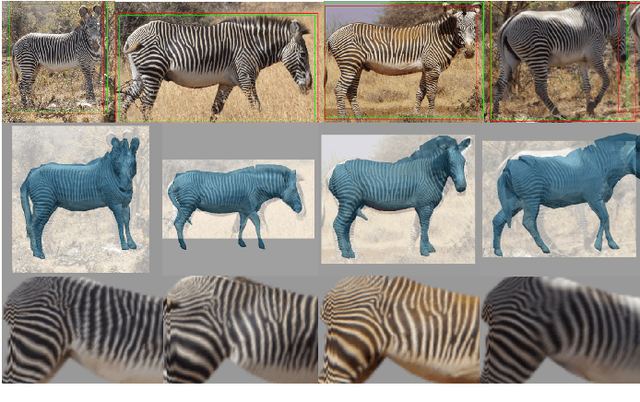
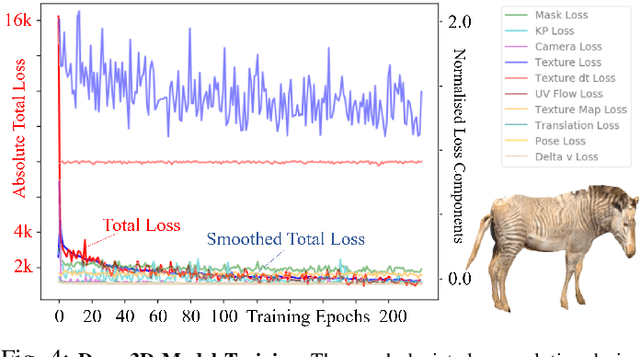
Abstract:This paper combines deep learning techniques for species detection, 3D model fitting, and metric learning in one pipeline to perform individual animal identification from photographs by exploiting unique coat patterns. This is the first work to attempt this and, compared to traditional 2D bounding box or segmentation based CNN identification pipelines, the approach provides effective and explicit view-point normalisation and allows for a straight forward visualisation of the learned biometric population space. Note that due to the use of metric learning the pipeline is also readily applicable to open set and zero shot re-identification scenarios. We apply the proposed approach to individual Grevy's zebra (Equus grevyi) identification and show in a small study on the SMALST dataset that the use of 3D model fitting can indeed benefit performance. In particular, back-projected textures from 3D fitted models improve identification accuracy from 48.0% to 56.8% compared to 2D bounding box approaches for the dataset. Whilst the study is far too small accurately to estimate the full performance potential achievable in larger-scale real-world application settings and in comparisons against polished tools, our work lays the conceptual and practical foundations for a next step in animal biometrics towards deep metric learning driven, fully 3D-aware animal identification in open population settings. We publish network weights and relevant facilitating source code with this paper for full reproducibility and as inspiration for further research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge