Susana K. Lai-Yuen
crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023
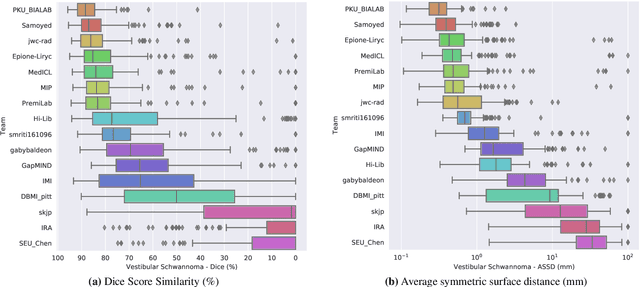
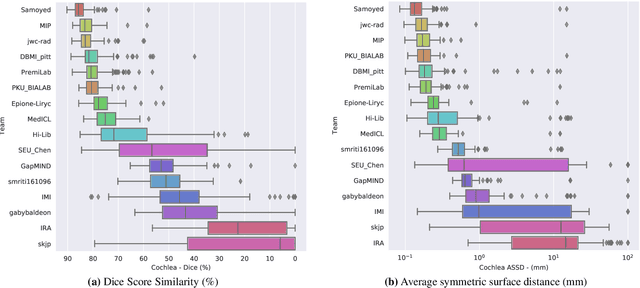
Jun 13, 2025Abstract:The cross-Modality Domain Adaptation (crossMoDA) challenge series, initiated in 2021 in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), focuses on unsupervised cross-modality segmentation, learning from contrast-enhanced T1 (ceT1) and transferring to T2 MRI. The task is an extreme example of domain shift chosen to serve as a meaningful and illustrative benchmark. From a clinical application perspective, it aims to automate Vestibular Schwannoma (VS) and cochlea segmentation on T2 scans for more cost-effective VS management. Over time, the challenge objectives have evolved to enhance its clinical relevance. The challenge evolved from using single-institutional data and basic segmentation in 2021 to incorporating multi-institutional data and Koos grading in 2022, and by 2023, it included heterogeneous routine data and sub-segmentation of intra- and extra-meatal tumour components. In this work, we report the findings of the 2022 and 2023 editions and perform a retrospective analysis of the challenge progression over the years. The observations from the successive challenge contributions indicate that the number of outliers decreases with an expanding dataset. This is notable since the diversity of scanning protocols of the datasets concurrently increased. The winning approach of the 2023 edition reduced the number of outliers on the 2021 and 2022 testing data, demonstrating how increased data heterogeneity can enhance segmentation performance even on homogeneous data. However, the cochlea Dice score declined in 2023, likely due to the added complexity from tumour sub-annotations affecting overall segmentation performance. While progress is still needed for clinically acceptable VS segmentation, the plateauing performance suggests that a more challenging cross-modal task may better serve future benchmarking.
CrossMoDA 2021 challenge: Benchmark of Cross-Modality Domain Adaptation techniques for Vestibular Schwnannoma and Cochlea Segmentation
Jan 08, 2022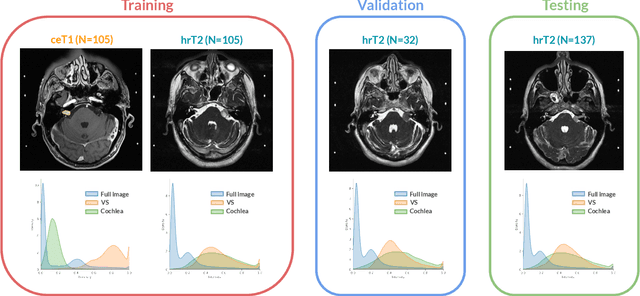
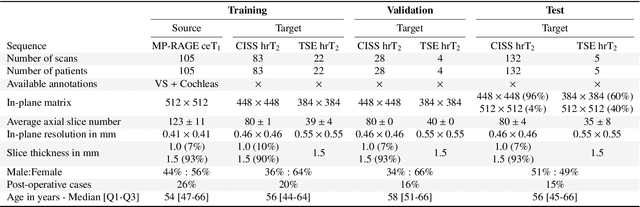
Abstract:Domain Adaptation (DA) has recently raised strong interests in the medical imaging community. While a large variety of DA techniques has been proposed for image segmentation, most of these techniques have been validated either on private datasets or on small publicly available datasets. Moreover, these datasets mostly addressed single-class problems. To tackle these limitations, the Cross-Modality Domain Adaptation (crossMoDA) challenge was organised in conjunction with the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2021). CrossMoDA is the first large and multi-class benchmark for unsupervised cross-modality DA. The challenge's goal is to segment two key brain structures involved in the follow-up and treatment planning of vestibular schwannoma (VS): the VS and the cochleas. Currently, the diagnosis and surveillance in patients with VS are performed using contrast-enhanced T1 (ceT1) MRI. However, there is growing interest in using non-contrast sequences such as high-resolution T2 (hrT2) MRI. Therefore, we created an unsupervised cross-modality segmentation benchmark. The training set provides annotated ceT1 (N=105) and unpaired non-annotated hrT2 (N=105). The aim was to automatically perform unilateral VS and bilateral cochlea segmentation on hrT2 as provided in the testing set (N=137). A total of 16 teams submitted their algorithm for the evaluation phase. The level of performance reached by the top-performing teams is strikingly high (best median Dice - VS:88.4%; Cochleas:85.7%) and close to full supervision (median Dice - VS:92.5%; Cochleas:87.7%). All top-performing methods made use of an image-to-image translation approach to transform the source-domain images into pseudo-target-domain images. A segmentation network was then trained using these generated images and the manual annotations provided for the source image.
C-MADA: Unsupervised Cross-Modality Adversarial Domain Adaptation framework for medical Image Segmentation
Oct 29, 2021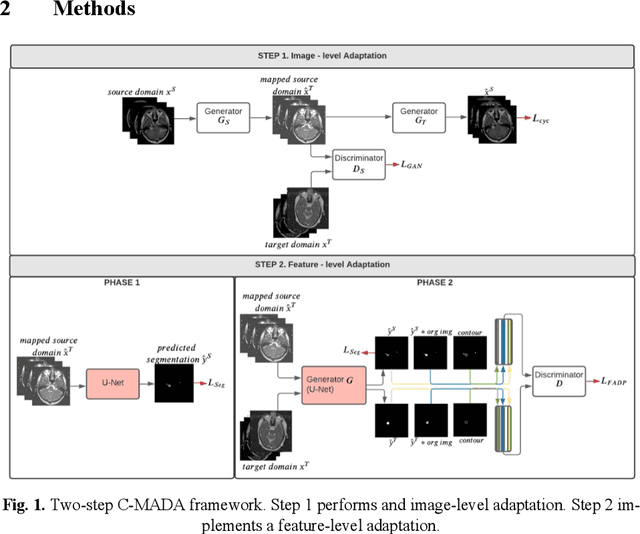
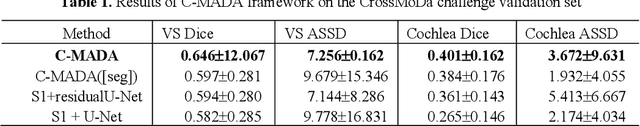
Abstract:Deep learning models have obtained state-of-the-art results for medical image analysis. However, when these models are tested on an unseen domain there is a significant performance degradation. In this work, we present an unsupervised Cross-Modality Adversarial Domain Adaptation (C-MADA) framework for medical image segmentation. C-MADA implements an image- and feature-level adaptation method in a sequential manner. First, images from the source domain are translated to the target domain through an un-paired image-to-image adversarial translation with cycle-consistency loss. Then, a U-Net network is trained with the mapped source domain images and target domain images in an adversarial manner to learn domain-invariant feature representations. Furthermore, to improve the networks segmentation performance, information about the shape, texture, and con-tour of the predicted segmentation is included during the adversarial train-ing. C-MADA is tested on the task of brain MRI segmentation, obtaining competitive results.
Self-Adaptive 2D-3D Ensemble of Fully Convolutional Networks for Medical Image Segmentation
Jul 26, 2019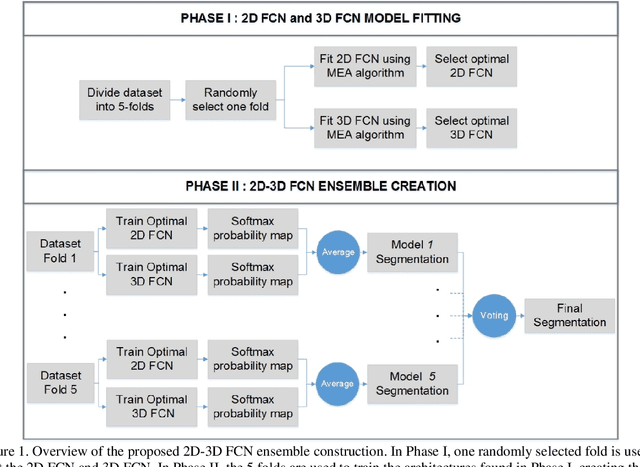
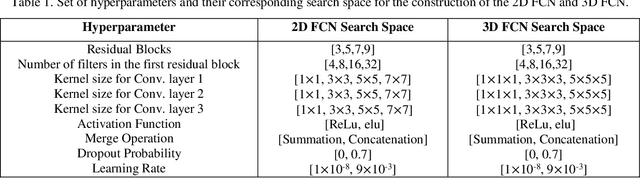
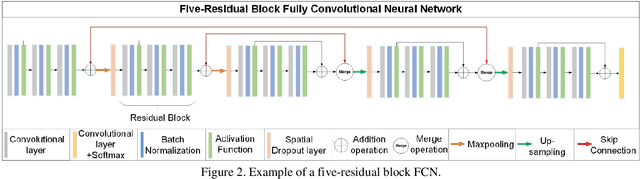
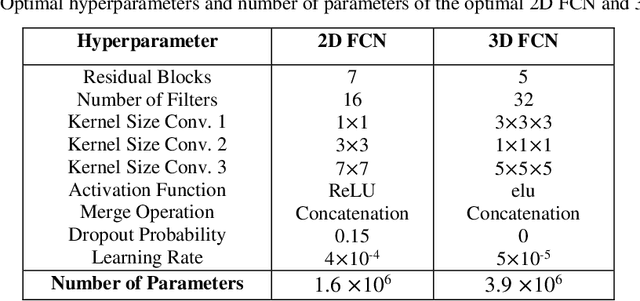
Abstract:Segmentation is a critical step in medical image analysis. Fully Convolutional Networks (FCNs) have emerged as powerful segmentation models achieving state-of-the-art results in various medical image datasets. Network architectures are usually designed manually for a specific segmentation task so applying them to other medical datasets requires extensive experience and time. Moreover, the segmentation requires handling large volumetric data that results in big and complex architectures. Recently, methods that automatically design neural networks for medical image segmentation have been presented; however, most approaches either do not fully consider volumetric information or do not optimize the size of the network. In this paper, we propose a novel self-adaptive 2D-3D ensemble of FCNs for medical image segmentation that incorporates volumetric information and optimizes both the model's performance and size. The model is composed of an ensemble of a 2D FCN that extracts intra-slice information, and a 3D FCN that exploits inter-slice information. The architectures of the 2D and 3D FCNs are automatically adapted to a medical image dataset using a multiobjective evolutionary based algorithm that minimizes both the segmentation error and number of parameters in the network. The proposed 2D-3D FCN ensemble was tested on the task of prostate segmentation on the image dataset from the PROMISE12 Grand Challenge. The resulting network is ranked in the top 10 submissions, surpassing the performance of other automatically-designed architectures while being considerably smaller in size.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge