Sixian You
System- and Sample-agnostic Isotropic 3D Microscopy by Weakly Physics-informed, Domain-shift-resistant Axial Deblurring
Jun 10, 2024



Abstract:Three-dimensional (3D) subcellular imaging is essential for biomedical research, but the diffraction limit of optical microscopy compromises axial resolution, hindering accurate 3D structural analysis. This challenge is particularly pronounced in label-free imaging of thick, heterogeneous tissues, where assumptions about data distribution (e.g. sparsity, label-specific distribution, and lateral-axial similarity) and system priors (e.g. independent and identically distributed (i.i.d.) noise and linear shift-invariant (LSI) point-spread functions (PSFs)) are often invalid. Here, we introduce SSAI-3D, a weakly physics-informed, domain-shift-resistant framework for robust isotropic 3D imaging. SSAI-3D enables robust axial deblurring by generating a PSF-flexible, noise-resilient, sample-informed training dataset and sparsely fine-tuning a large pre-trained blind deblurring network. SSAI-3D was applied to label-free nonlinear imaging of living organoids, freshly excised human endometrium tissue, and mouse whisker pads, and further validated in publicly available ground-truth-paired experimental datasets of 3D heterogeneous biological tissues with unknown blurring and noise across different microscopy systems.
Learned, Uncertainty-driven Adaptive Acquisition for Photon-Efficient Multiphoton Microscopy
Oct 24, 2023Abstract:Multiphoton microscopy (MPM) is a powerful imaging tool that has been a critical enabler for live tissue imaging. However, since most multiphoton microscopy platforms rely on point scanning, there is an inherent trade-off between acquisition time, field of view (FOV), phototoxicity, and image quality, often resulting in noisy measurements when fast, large FOV, and/or gentle imaging is needed. Deep learning could be used to denoise multiphoton microscopy measurements, but these algorithms can be prone to hallucination, which can be disastrous for medical and scientific applications. We propose a method to simultaneously denoise and predict pixel-wise uncertainty for multiphoton imaging measurements, improving algorithm trustworthiness and providing statistical guarantees for the deep learning predictions. Furthermore, we propose to leverage this learned, pixel-wise uncertainty to drive an adaptive acquisition technique that rescans only the most uncertain regions of a sample. We demonstrate our method on experimental noisy MPM measurements of human endometrium tissues, showing that we can maintain fine features and outperform other denoising methods while predicting uncertainty at each pixel. Finally, with our adaptive acquisition technique, we demonstrate a 120X reduction in acquisition time and total light dose while successfully recovering fine features in the sample. We are the first to demonstrate distribution-free uncertainty quantification for a denoising task with real experimental data and the first to propose adaptive acquisition based on reconstruction uncertainty
EventLFM: Event Camera integrated Fourier Light Field Microscopy for Ultrafast 3D imaging
Oct 01, 2023Abstract:Ultrafast 3D imaging is indispensable for visualizing complex and dynamic biological processes. Conventional scanning-based techniques necessitate an inherent tradeoff between the acquisition speed and space-bandwidth product (SBP). While single-shot 3D wide-field techniques have emerged as an attractive solution, they are still bottlenecked by the synchronous readout constraints of conventional CMOS architectures, thereby limiting the data throughput by frame rate to maintain a high SBP. Here, we present EventLFM, a straightforward and cost-effective system that circumnavigates these challenges by integrating an event camera with Fourier light field microscopy (LFM), a single-shot 3D wide-field imaging technique. The event camera operates on a novel asynchronous readout architecture, thereby bypassing the frame rate limitations intrinsic to conventional CMOS systems. We further develop a simple and robust event-driven LFM reconstruction algorithm that can reliably reconstruct 3D dynamics from the unique spatiotemporal measurements from EventLFM. We experimentally demonstrate that EventLFM can robustly image fast-moving and rapidly blinking 3D samples at KHz frame rates and furthermore, showcase EventLFM's ability to achieve 3D tracking of GFP-labeled neurons in freely moving C. elegans. We believe that the combined ultrafast speed and large 3D SBP offered by EventLFM may open up new possibilities across many biomedical applications.
Roadmap on Deep Learning for Microscopy
Mar 07, 2023



Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Linear Revolution-Invariance: Modeling and Deblurring Spatially-Varying Imaging Systems
Jun 17, 2022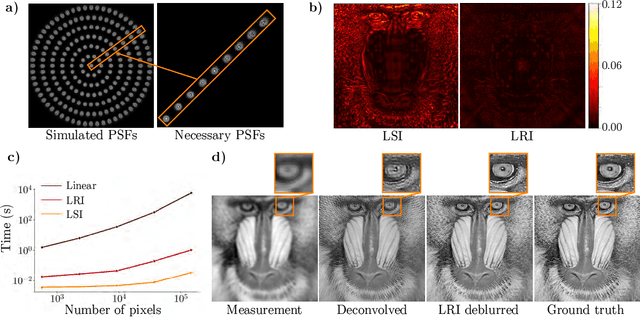
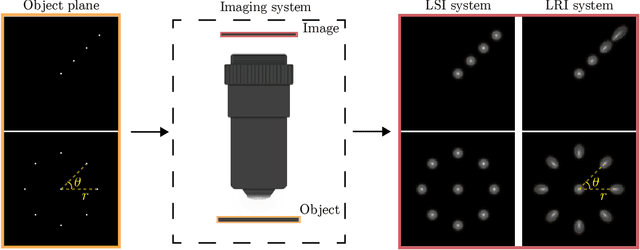
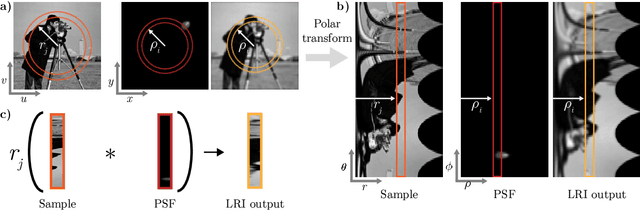
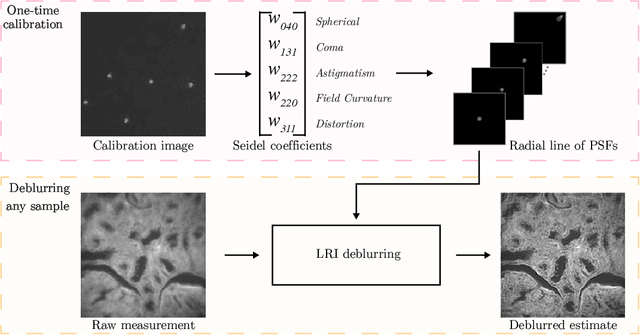
Abstract:We develop theory and algorithms for modeling and deblurring imaging systems that are composed of rotationally-symmetric optics. Such systems have point spread functions (PSFs) which are spatially-varying, but only vary radially, a property we call linear revolution-invariance (LRI). From the LRI property we develop an exact theory for linear imaging with radially-varying optics, including an analog of the Fourier Convolution Theorem. This theory, in tandem with a calibration procedure using Seidel aberration coefficients, yields an efficient forward model and deblurring algorithm which requires only a single calibration image -- one that is easier to measure than a single PSF. We test these methods in simulation and experimentally on images of resolution targets, rabbit liver tissue, and live tardigrades obtained using the UCLA Miniscope v3. We find that the LRI forward model generates accurate radially-varying blur, and LRI deblurring improves resolution, especially near the edges of the field-of-view. These methods are available for use as a Python package at https://github.com/apsk14/lri.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge