Sharib Ali
An objective validation of polyp and instrument segmentation methods in colonoscopy through Medico 2020 polyp segmentation and MedAI 2021 transparency challenges
Jul 30, 2023



Abstract:Automatic analysis of colonoscopy images has been an active field of research motivated by the importance of early detection of precancerous polyps. However, detecting polyps during the live examination can be challenging due to various factors such as variation of skills and experience among the endoscopists, lack of attentiveness, and fatigue leading to a high polyp miss-rate. Deep learning has emerged as a promising solution to this challenge as it can assist endoscopists in detecting and classifying overlooked polyps and abnormalities in real time. In addition to the algorithm's accuracy, transparency and interpretability are crucial to explaining the whys and hows of the algorithm's prediction. Further, most algorithms are developed in private data, closed source, or proprietary software, and methods lack reproducibility. Therefore, to promote the development of efficient and transparent methods, we have organized the "Medico automatic polyp segmentation (Medico 2020)" and "MedAI: Transparency in Medical Image Segmentation (MedAI 2021)" competitions. We present a comprehensive summary and analyze each contribution, highlight the strength of the best-performing methods, and discuss the possibility of clinical translations of such methods into the clinic. For the transparency task, a multi-disciplinary team, including expert gastroenterologists, accessed each submission and evaluated the team based on open-source practices, failure case analysis, ablation studies, usability and understandability of evaluations to gain a deeper understanding of the models' credibility for clinical deployment. Through the comprehensive analysis of the challenge, we not only highlight the advancements in polyp and surgical instrument segmentation but also encourage qualitative evaluation for building more transparent and understandable AI-based colonoscopy systems.
A Client-server Deep Federated Learning for Cross-domain Surgical Image Segmentation
Jun 14, 2023



Abstract:This paper presents a solution to the cross-domain adaptation problem for 2D surgical image segmentation, explicitly considering the privacy protection of distributed datasets belonging to different centers. Deep learning architectures in medical image analysis necessitate extensive training data for better generalization. However, obtaining sufficient diagnostic and surgical data is still challenging, mainly due to the inherent cost of data curation and the need of experts for data annotation. Moreover, increased privacy and legal compliance concerns can make data sharing across clinical sites or regions difficult. Another ubiquitous challenge the medical datasets face is inevitable domain shifts among the collected data at the different centers. To this end, we propose a Client-server deep federated architecture for cross-domain adaptation. A server hosts a set of immutable parameters common to both the source and target domains. The clients consist of the respective domain-specific parameters and make requests to the server while learning their parameters and inferencing. We evaluate our framework in two benchmark datasets, demonstrating applicability in computer-assisted interventions for endoscopic polyp segmentation and diagnostic skin lesion detection and analysis. Our extensive quantitative and qualitative experiments demonstrate the superiority of the proposed method compared to competitive baseline and state-of-the-art methods. Codes are available at: https://github.com/thetna/distributed-da
SSL-CPCD: Self-supervised learning with composite pretext-class discrimination for improved generalisability in endoscopic image analysis
May 31, 2023Abstract:Data-driven methods have shown tremendous progress in medical image analysis. In this context, deep learning-based supervised methods are widely popular. However, they require a large amount of training data and face issues in generalisability to unseen datasets that hinder clinical translation. Endoscopic imaging data incorporates large inter- and intra-patient variability that makes these models more challenging to learn representative features for downstream tasks. Thus, despite the publicly available datasets and datasets that can be generated within hospitals, most supervised models still underperform. While self-supervised learning has addressed this problem to some extent in natural scene data, there is a considerable performance gap in the medical image domain. In this paper, we propose to explore patch-level instance-group discrimination and penalisation of inter-class variation using additive angular margin within the cosine similarity metrics. Our novel approach enables models to learn to cluster similar representative patches, thereby improving their ability to provide better separation between different classes. Our results demonstrate significant improvement on all metrics over the state-of-the-art (SOTA) methods on the test set from the same and diverse datasets. We evaluated our approach for classification, detection, and segmentation. SSL-CPCD achieves 79.77% on Top 1 accuracy for ulcerative colitis classification, 88.62% on mAP for polyp detection, and 82.32% on dice similarity coefficient for segmentation tasks are nearly over 4%, 2%, and 3%, respectively, compared to the baseline architectures. We also demonstrate that our method generalises better than all SOTA methods to unseen datasets, reporting nearly 7% improvement in our generalisability assessment.
Why is the winner the best?
Mar 30, 2023



Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
SUPRA: Superpixel Guided Loss for Improved Multi-modal Segmentation in Endoscopy
Nov 12, 2022



Abstract:Domain shift is a well-known problem in the medical imaging community. In particular, for endoscopic image analysis where the data can have different modalities the performance of deep learning (DL) methods gets adversely affected. In other words, methods developed on one modality cannot be used for a different modality. However, in real clinical settings, endoscopists switch between modalities for better mucosal visualisation. In this paper, we explore the domain generalisation technique to enable DL methods to be used in such scenarios. To this extend, we propose to use super pixels generated with Simple Linear Iterative Clustering (SLIC) which we refer to as "SUPRA" for SUPeRpixel Augmented method. SUPRA first generates a preliminary segmentation mask making use of our new loss "SLICLoss" that encourages both an accurate and color-consistent segmentation. We demonstrate that SLICLoss when combined with Binary Cross Entropy loss (BCE) can improve the model's generalisability with data that presents significant domain shift. We validate this novel compound loss on a vanilla U-Net using the EndoUDA dataset, which contains images for Barret's Esophagus and polyps from two modalities. We show that our method yields an improvement of nearly 25% in the target domain set compared to the baseline.
A comprehensive survey on recent deep learning-based methods applied to surgical data
Sep 03, 2022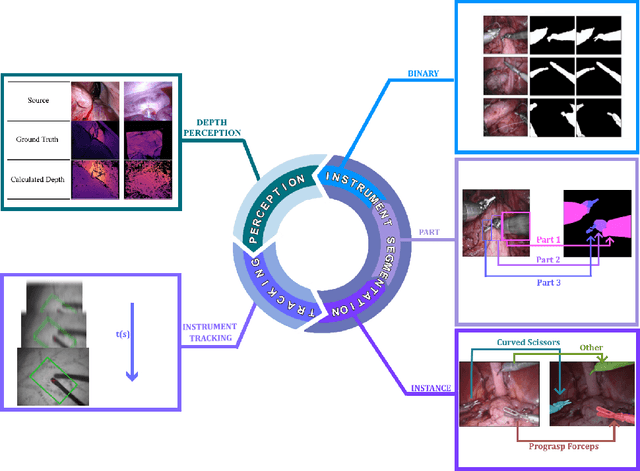
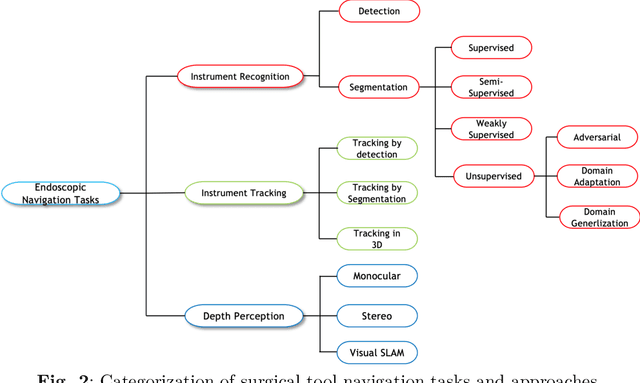
Abstract:Minimally invasive surgery is highly operator dependant with lengthy procedural times causing fatigue and risk to patients. In order to mitigate these risks, real-time systems can help assist surgeons to navigate and track tools, by providing clear understanding of scene and avoid miscalculations during operation. While several efforts have been made in this direction, a lack of diverse datasets, as well as very dynamic scenes and its variability in each patient entails major hurdle in accomplishing robust systems. In this work, we present a systematic review of recent machine learning-based approaches including surgical tool localisation, segmentation, tracking and 3D scene perception. Furthermore, we present current gaps and directions of these invented methods and provide rational behind clinical integration of these approaches.
A semi-supervised Teacher-Student framework for surgical tool detection and localization
Aug 21, 2022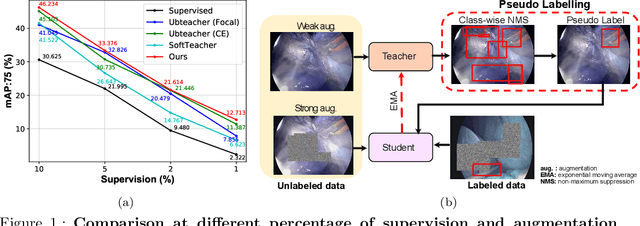
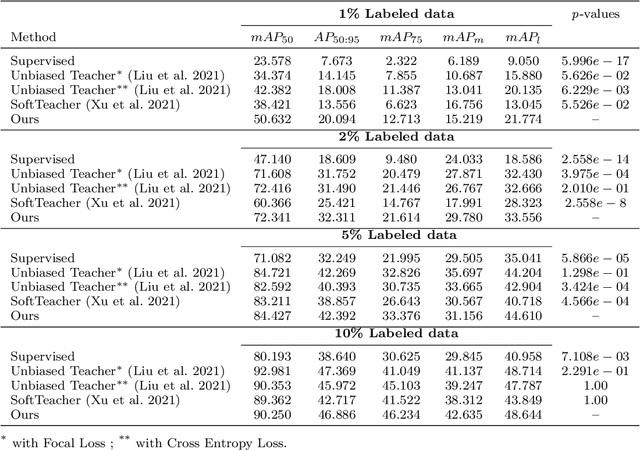
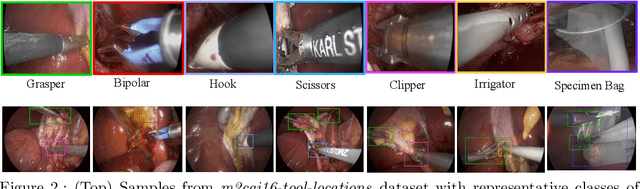
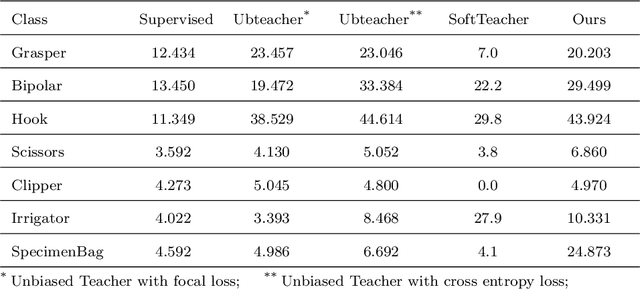
Abstract:Surgical tool detection in minimally invasive surgery is an essential part of computer-assisted interventions. Current approaches are mostly based on supervised methods which require large fully labeled data to train supervised models and suffer from pseudo label bias because of class imbalance issues. However large image datasets with bounding box annotations are often scarcely available. Semi-supervised learning (SSL) has recently emerged as a means for training large models using only a modest amount of annotated data; apart from reducing the annotation cost. SSL has also shown promise to produce models that are more robust and generalizable. Therefore, in this paper we introduce a semi-supervised learning (SSL) framework in surgical tool detection paradigm which aims to mitigate the scarcity of training data and the data imbalance through a knowledge distillation approach. In the proposed work, we train a model with labeled data which initialises the Teacher-Student joint learning, where the Student is trained on Teacher-generated pseudo labels from unlabeled data. We propose a multi-class distance with a margin based classification loss function in the region-of-interest head of the detector to effectively segregate foreground classes from background region. Our results on m2cai16-tool-locations dataset indicate the superiority of our approach on different supervised data settings (1%, 2%, 5%, 10% of annotated data) where our model achieves overall improvements of 8%, 12% and 27% in mAP (on 1% labeled data) over the state-of-the-art SSL methods and a fully supervised baseline, respectively. The code is available at https://github.com/Mansoor-at/Semi-supervised-surgical-tool-det
Knowledge distillation with a class-aware loss for endoscopic disease detection
Jul 19, 2022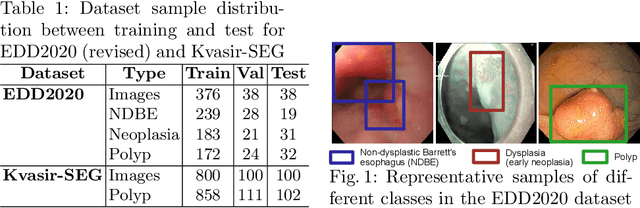
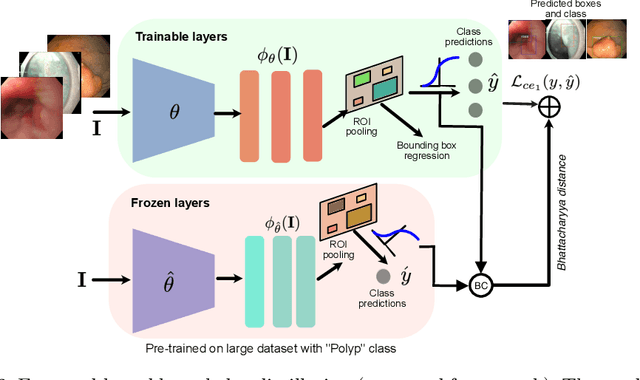
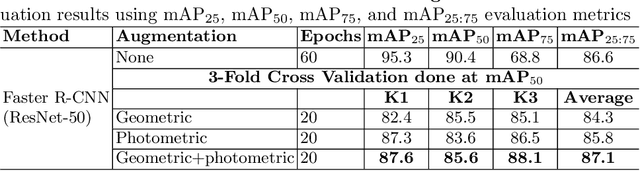
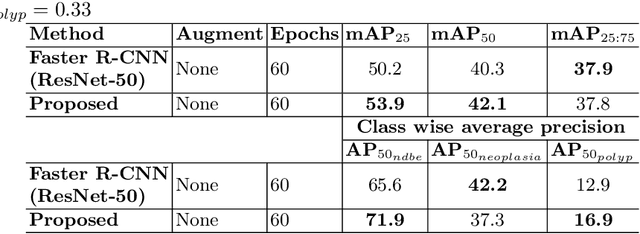
Abstract:Prevalence of gastrointestinal (GI) cancer is growing alarmingly every year leading to a substantial increase in the mortality rate. Endoscopic detection is providing crucial diagnostic support, however, subtle lesions in upper and lower GI are quite hard to detect and cause considerable missed detection. In this work, we leverage deep learning to develop a framework to improve the localization of difficult to detect lesions and minimize the missed detection rate. We propose an end to end student-teacher learning setup where class probabilities of a trained teacher model on one class with larger dataset are used to penalize multi-class student network. Our model achieves higher performance in terms of mean average precision (mAP) on both endoscopic disease detection (EDD2020) challenge and Kvasir-SEG datasets. Additionally, we show that using such learning paradigm, our model is generalizable to unseen test set giving higher APs for clinically crucial neoplastic and polyp categories
Patch-level instance-group discrimination with pretext-invariant learning for colitis scoring
Jul 11, 2022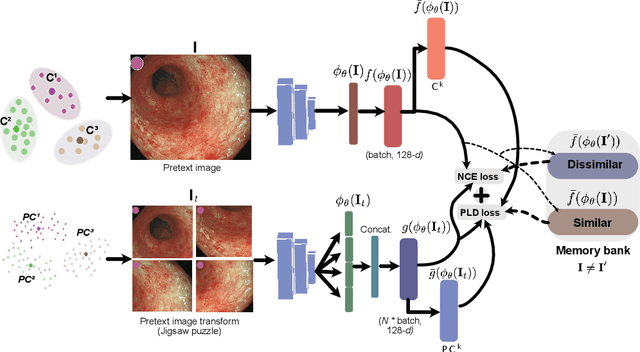
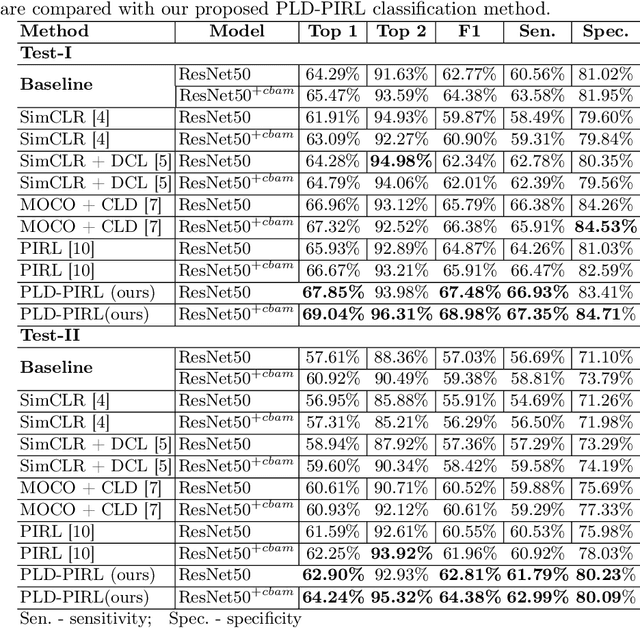
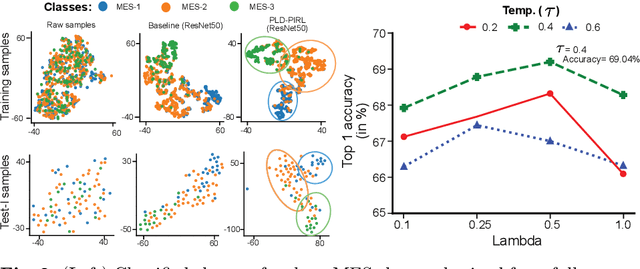
Abstract:Inflammatory bowel disease (IBD), in particular ulcerative colitis (UC), is graded by endoscopists and this assessment is the basis for risk stratification and therapy monitoring. Presently, endoscopic characterisation is largely operator dependant leading to sometimes undesirable clinical outcomes for patients with IBD. We focus on the Mayo Endoscopic Scoring (MES) system which is widely used but requires the reliable identification of subtle changes in mucosal inflammation. Most existing deep learning classification methods cannot detect these fine-grained changes which make UC grading such a challenging task. In this work, we introduce a novel patch-level instance-group discrimination with pretext-invariant representation learning (PLD-PIRL) for self-supervised learning (SSL). Our experiments demonstrate both improved accuracy and robustness compared to the baseline supervised network and several state-of-the-art SSL methods. Compared to the baseline (ResNet50) supervised classification our proposed PLD-PIRL obtained an improvement of 4.75% on hold-out test data and 6.64% on unseen center test data for top-1 accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge