Shadi Albarqouni
University Hospital Bonn, Venusberg-Campus 1, D-53127, Bonn, Germany, Helmholtz Munich, Ingolstädter Landstraße 1, D-85764, Neuherberg, Germany, Technical University of Munich, Boltzmannstr. 3, D-85748 Garching, Germany
Sickle Cell Disease Severity Prediction from Percoll Gradient Images using Graph Convolutional Networks
Sep 11, 2021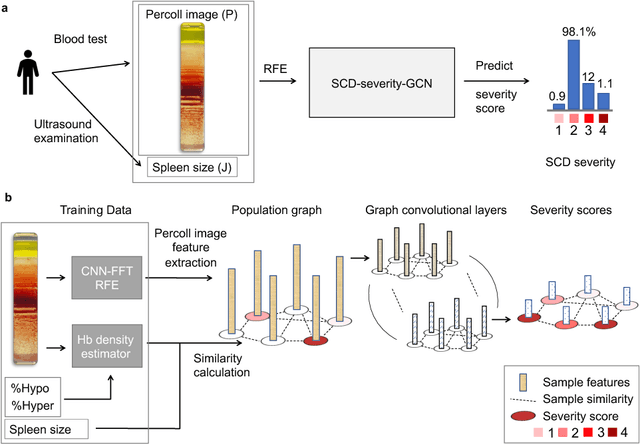
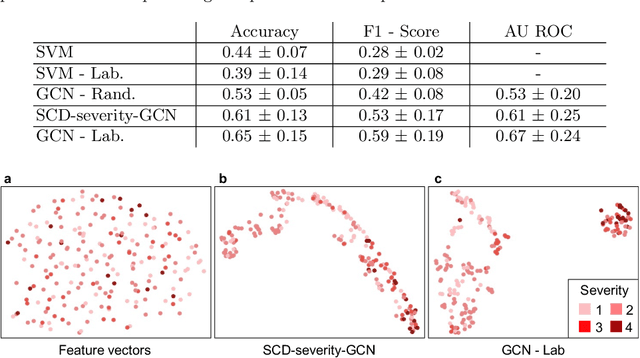
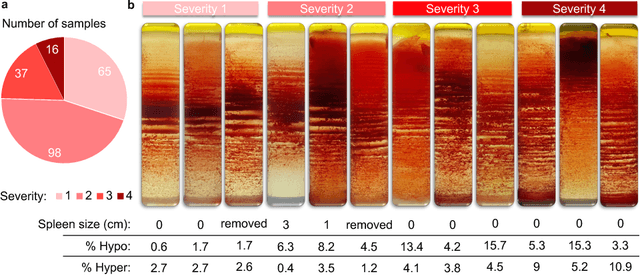
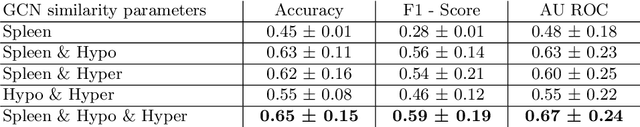
Abstract:Sickle cell disease (SCD) is a severe genetic hemoglobin disorder that results in premature destruction of red blood cells. Assessment of the severity of the disease is a challenging task in clinical routine since the causes of broad variance in SCD manifestation despite the common genetic cause remain unclear. Identification of the biomarkers that would predict the severity grade is of importance for prognosis and assessment of responsiveness of patients to therapy. Detection of the changes in red blood cell (RBC) density through separation of Percoll density gradient could be such marker as it allows to resolve intercellular differences and follow the most damaged dense cells prone to destruction and vaso-occlusion. Quantification of the images obtained from the distribution of RBCs in Percoll gradient and interpretation of the obtained is an important prerequisite for establishment of this approach. Here, we propose a novel approach combining a graph convolutional network, a convolutional neural network, fast Fourier transform, and recursive feature elimination to predict the severity of SCD directly from a Percoll image. Two important but expensive laboratory blood test parameters measurements are used for training the graph convolutional network. To make the model independent from such tests during prediction, the two parameters are estimated by a neural network from the Percoll image directly. On a cohort of 216 subjects, we achieve a prediction performance that is only slightly below an approach where the groundtruth laboratory measurements are used. Our proposed method is the first computational approach for the difficult task of SCD severity prediction. The two-step approach relies solely on inexpensive and simple blood analysis tools and can have a significant impact on the patients' survival in underdeveloped countries where access to medical instruments and doctors is limited
The Federated Tumor Segmentation (FeTS) Challenge
May 14, 2021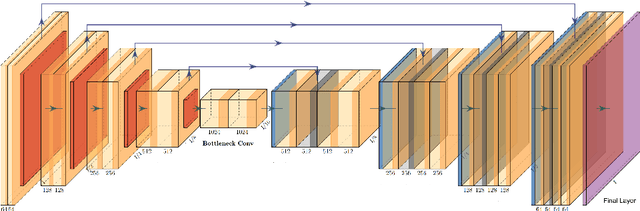
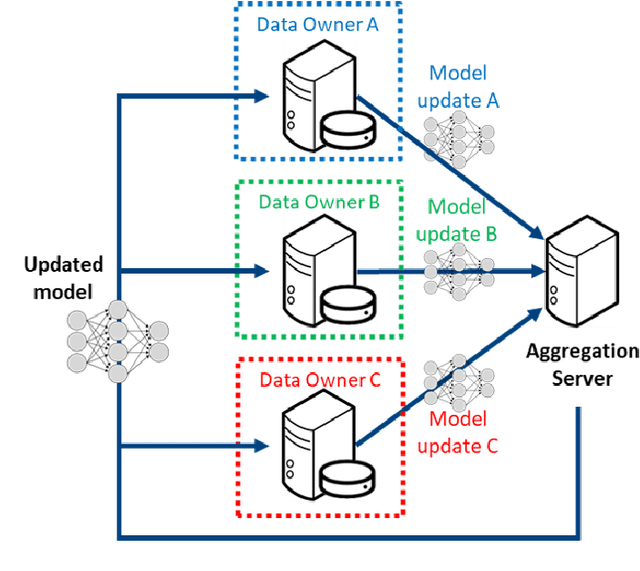
Abstract:This manuscript describes the first challenge on Federated Learning, namely the Federated Tumor Segmentation (FeTS) challenge 2021. International challenges have become the standard for validation of biomedical image analysis methods. However, the actual performance of participating (even the winning) algorithms on "real-world" clinical data often remains unclear, as the data included in challenges are usually acquired in very controlled settings at few institutions. The seemingly obvious solution of just collecting increasingly more data from more institutions in such challenges does not scale well due to privacy and ownership hurdles. Towards alleviating these concerns, we are proposing the FeTS challenge 2021 to cater towards both the development and the evaluation of models for the segmentation of intrinsically heterogeneous (in appearance, shape, and histology) brain tumors, namely gliomas. Specifically, the FeTS 2021 challenge uses clinically acquired, multi-institutional magnetic resonance imaging (MRI) scans from the BraTS 2020 challenge, as well as from various remote independent institutions included in the collaborative network of a real-world federation (https://www.fets.ai/). The goals of the FeTS challenge are directly represented by the two included tasks: 1) the identification of the optimal weight aggregation approach towards the training of a consensus model that has gained knowledge via federated learning from multiple geographically distinct institutions, while their data are always retained within each institution, and 2) the federated evaluation of the generalizability of brain tumor segmentation models "in the wild", i.e. on data from institutional distributions that were not part of the training datasets.
Fourier Transform of Percoll Gradients Boosts CNN Classification of Hereditary Hemolytic Anemias
Mar 17, 2021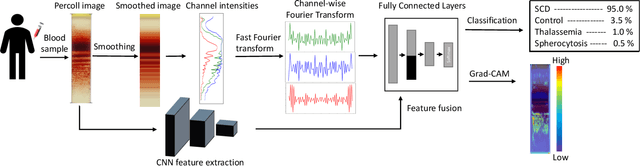
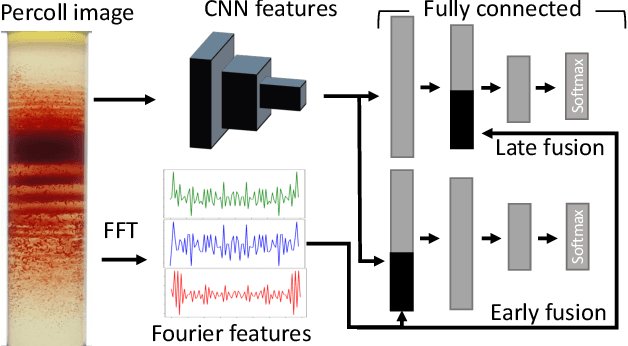
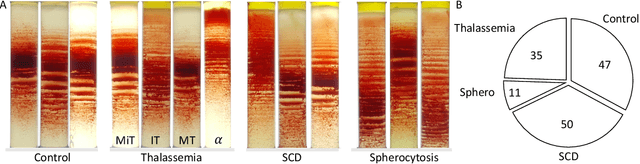
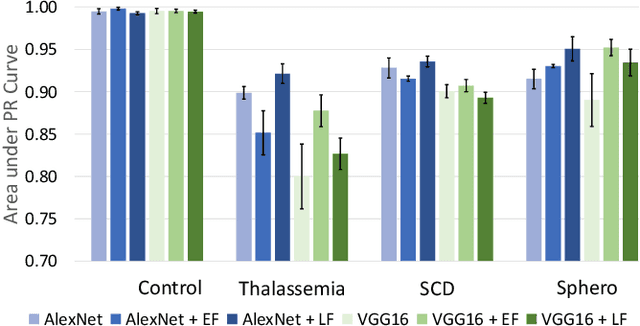
Abstract:Hereditary hemolytic anemias are genetic disorders that affect the shape and density of red blood cells. Genetic tests currently used to diagnose such anemias are expensive and unavailable in the majority of clinical labs. Here, we propose a method for identifying hereditary hemolytic anemias based on a standard biochemistry method, called Percoll gradient, obtained by centrifuging a patient's blood. Our hybrid approach consists on using spatial data-driven features, extracted with a convolutional neural network and spectral handcrafted features obtained from fast Fourier transform. We compare late and early feature fusion with AlexNet and VGG16 architectures. AlexNet with late fusion of spectral features performs better compared to other approaches. We achieved an average F1-score of 88% on different classes suggesting the possibility of diagnosing of hereditary hemolytic anemias from Percoll gradients. Finally, we utilize Grad-CAM to explore the spatial features used for classification.
Peer Learning for Skin Lesion Classification
Mar 08, 2021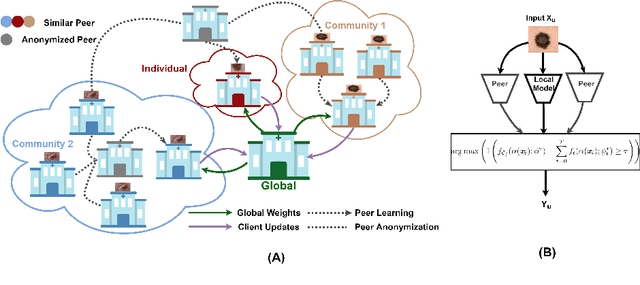
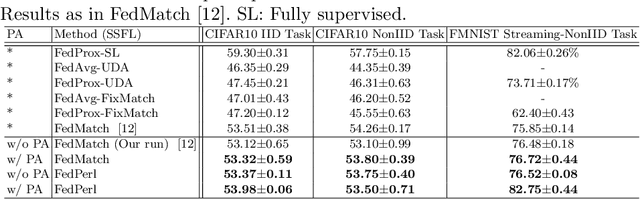
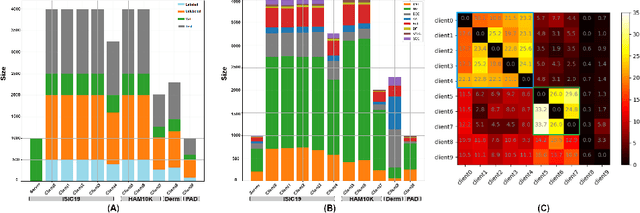
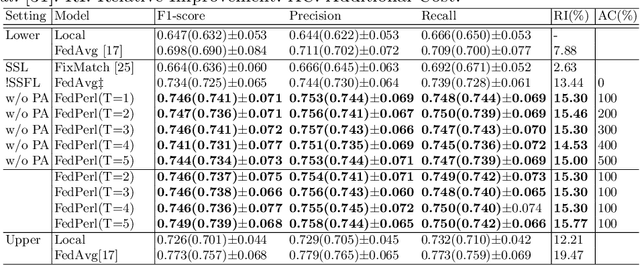
Abstract:Skin cancer is one of the most deadly cancers worldwide. Yet, it can be reduced by early detection. Recent deep-learning methods have shown a dermatologist-level performance in skin cancer classification. Yet, this success demands a large amount of centralized data, which is oftentimes not available. Federated learning has been recently introduced to train machine learning models in a privacy-preserved distributed fashion demanding annotated data at the clients, which is usually expensive and not available, especially in the medical field. To this end, we propose FedPerl, a semi-supervised federated learning method that utilizes peer learning from social sciences and ensemble averaging from committee machines to build communities and encourage its members to learn from each other such that they produce more accurate pseudo labels. We also propose the peer anonymization (PA) technique as a core component of FedPerl. PA preserves privacy and reduces the communication cost while maintaining the performance without additional complexity. We validated our method on 38,000 skin lesion images collected from 4 publicly available datasets. FedPerl achieves superior performance over the baselines and state-of-the-art SSFL by 15.8%, and 1.8% respectively. Further, FedPerl shows less sensitivity to noisy clients.
FedDis: Disentangled Federated Learning for Unsupervised Brain Pathology Segmentation
Mar 05, 2021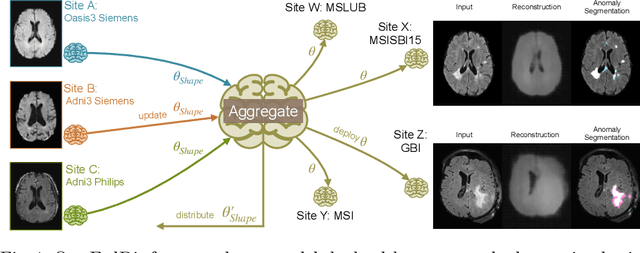
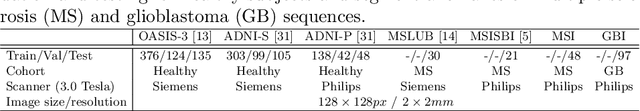
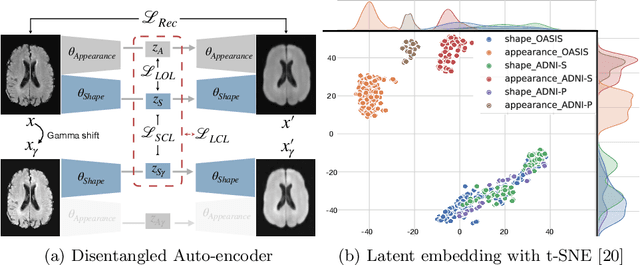
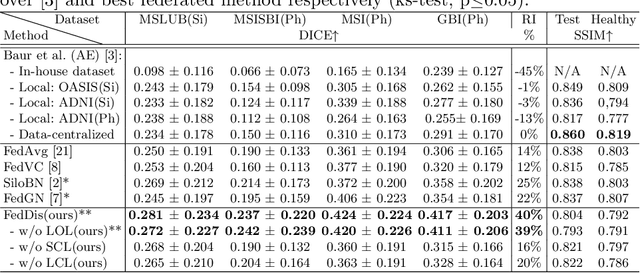
Abstract:In recent years, data-driven machine learning (ML) methods have revolutionized the computer vision community by providing novel efficient solutions to many unsolved (medical) image analysis problems. However, due to the increasing privacy concerns and data fragmentation on many different sites, existing medical data are not fully utilized, thus limiting the potential of ML. Federated learning (FL) enables multiple parties to collaboratively train a ML model without exchanging local data. However, data heterogeneity (non-IID) among the distributed clients is yet a challenge. To this end, we propose a novel federated method, denoted Federated Disentanglement (FedDis), to disentangle the parameter space into shape and appearance, and only share the shape parameter with the clients. FedDis is based on the assumption that the anatomical structure in brain MRI images is similar across multiple institutions, and sharing the shape knowledge would be beneficial in anomaly detection. In this paper, we leverage healthy brain scans of 623 subjects from multiple sites with real data (OASIS, ADNI) in a privacy-preserving fashion to learn a model of normal anatomy, that allows to segment abnormal structures. We demonstrate a superior performance of FedDis on real pathological databases containing 109 subjects; two publicly available MS Lesions (MSLUB, MSISBI), and an in-house database with MS and Glioblastoma (MSI and GBI). FedDis achieved an average dice performance of 0.38, outperforming the state-of-the-art (SOTA) auto-encoder by 42% and the SOTA federated method by 11%. Further, we illustrate that FedDis learns a shape embedding that is orthogonal to the appearance and consistent under different intensity augmentations.
An Uncertainty-Driven GCN Refinement Strategy for Organ Segmentation
Dec 06, 2020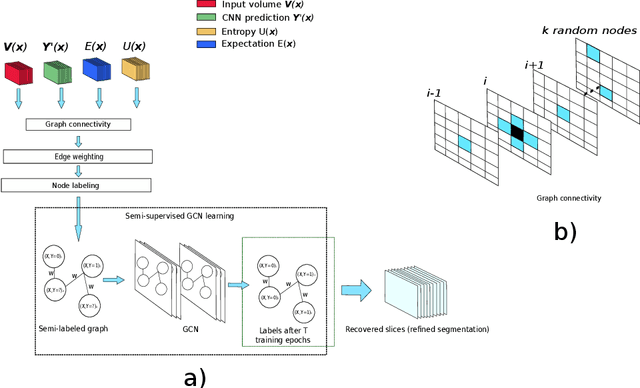
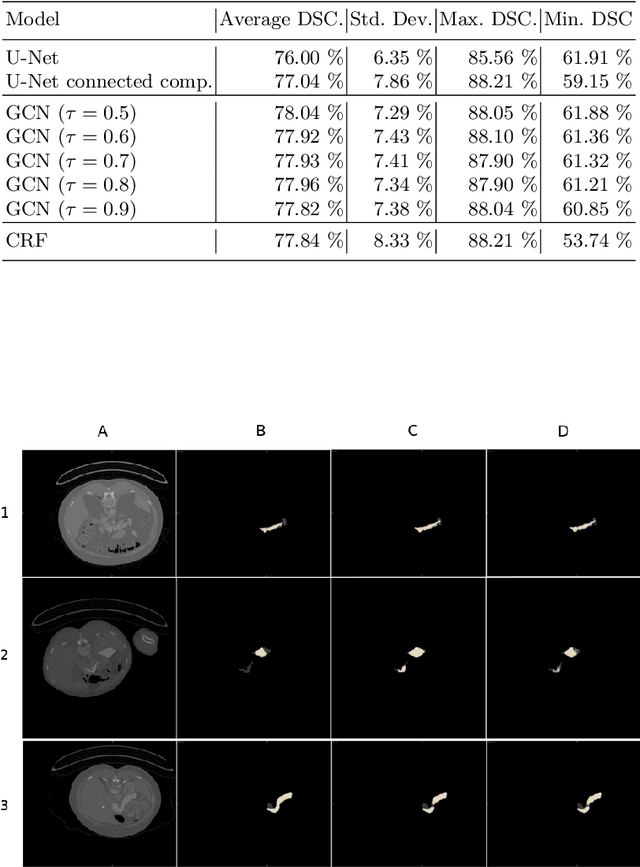
Abstract:Organ segmentation in CT volumes is an important pre-processing step in many computer assisted intervention and diagnosis methods. In recent years, convolutional neural networks have dominated the state of the art in this task. However, since this problem presents a challenging environment due to high variability in the organ's shape and similarity between tissues, the generation of false negative and false positive regions in the output segmentation is a common issue. Recent works have shown that the uncertainty analysis of the model can provide us with useful information about potential errors in the segmentation. In this context, we proposed a segmentation refinement method based on uncertainty analysis and graph convolutional networks. We employ the uncertainty levels of the convolutional network in a particular input volume to formulate a semi-supervised graph learning problem that is solved by training a graph convolutional network. To test our method we refine the initial output of a 2D U-Net. We validate our framework with the NIH pancreas dataset and the spleen dataset of the medical segmentation decathlon. We show that our method outperforms the state-of-the-art CRF refinement method by improving the dice score by 1% for the pancreas and 2% for spleen, with respect to the original U-Net's prediction. Finally, we perform a sensitivity analysis on the parameters of our proposal and discuss the applicability to other CNN architectures, the results, and current limitations of the model for future work in this research direction. For reproducibility purposes, we make our code publicly available at https://github.com/rodsom22/gcn_refinement.
Polyp-artifact relationship analysis using graph inductive learned representations
Sep 15, 2020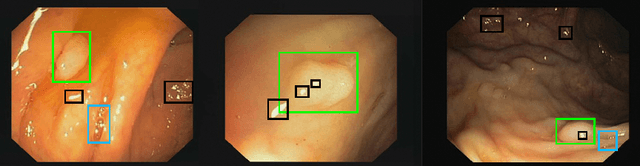
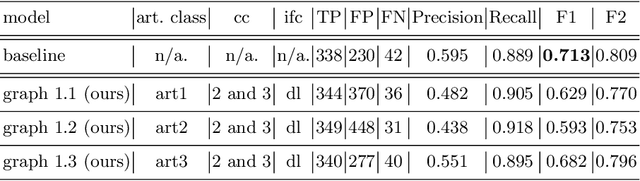

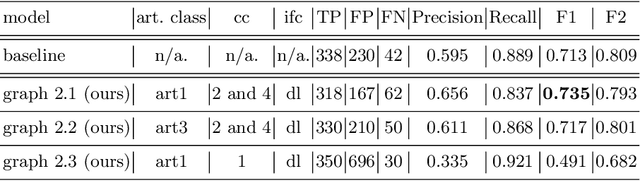
Abstract:The diagnosis process of colorectal cancer mainly focuses on the localization and characterization of abnormal growths in the colon tissue known as polyps. Despite recent advances in deep object localization, the localization of polyps remains challenging due to the similarities between tissues, and the high level of artifacts. Recent studies have shown the negative impact of the presence of artifacts in the polyp detection task, and have started to take them into account within the training process. However, the use of prior knowledge related to the spatial interaction of polyps and artifacts has not yet been considered. In this work, we incorporate artifact knowledge in a post-processing step. Our method models this task as an inductive graph representation learning problem, and is composed of training and inference steps. Detected bounding boxes around polyps and artifacts are considered as nodes connected by a defined criterion. The training step generates a node classifier with ground truth bounding boxes. In inference, we use this classifier to analyze a second graph, generated from artifact and polyp predictions given by region proposal networks. We evaluate how the choices in the connectivity and artifacts affect the performance of our method and show that it has the potential to reduce the false positives in the results of a region proposal network.
Inverse Distance Aggregation for Federated Learning with Non-IID Data
Aug 17, 2020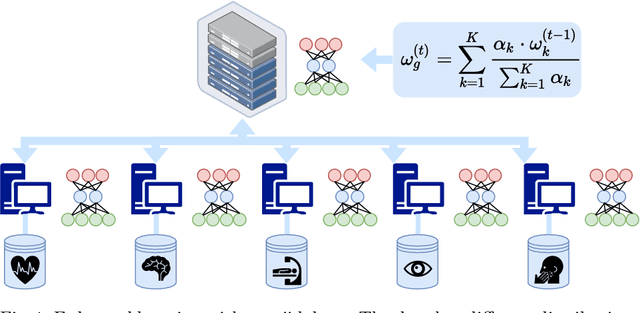
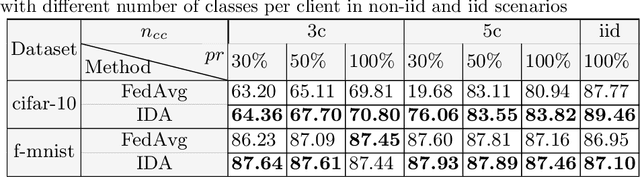
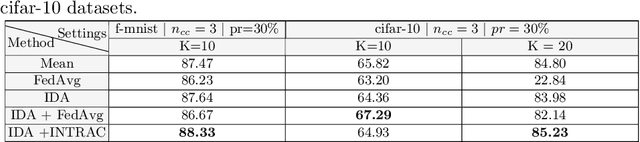
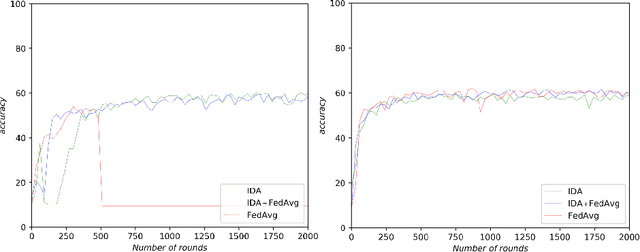
Abstract:Federated learning (FL) has been a promising approach in the field of medical imaging in recent years. A critical problem in FL, specifically in medical scenarios is to have a more accurate shared model which is robust to noisy and out-of distribution clients. In this work, we tackle the problem of statistical heterogeneity in data for FL which is highly plausible in medical data where for example the data comes from different sites with different scanner settings. We propose IDA (Inverse Distance Aggregation), a novel adaptive weighting approach for clients based on meta-information which handles unbalanced and non-iid data. We extensively analyze and evaluate our method against the well-known FL approach, Federated Averaging as a baseline.
Attention based Multiple Instance Learning for Classification of Blood Cell Disorders
Jul 22, 2020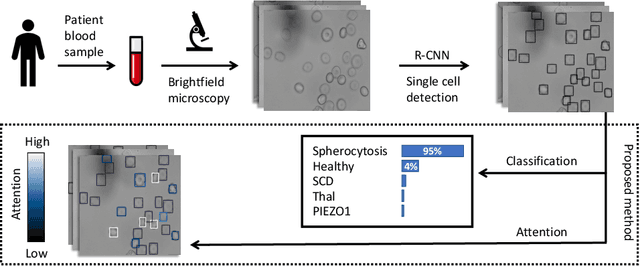
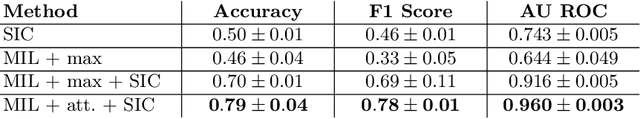
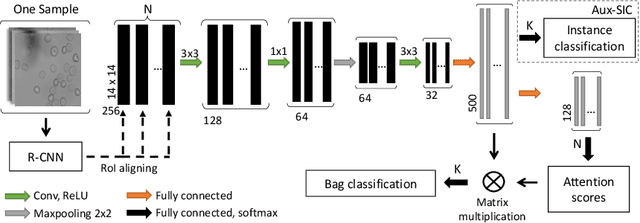
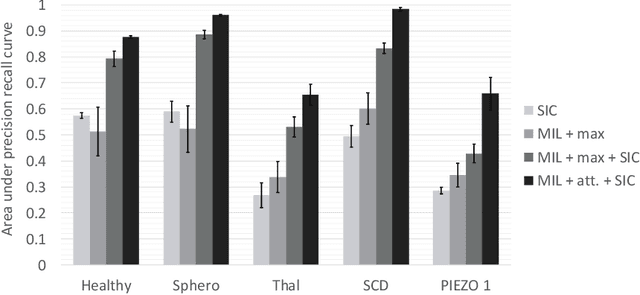
Abstract:Red blood cells are highly deformable and present in various shapes. In blood cell disorders, only a subset of all cells is morphologically altered and relevant for the diagnosis. However, manually labeling of all cells is laborious, complicated and introduces inter-expert variability. We propose an attention based multiple instance learning method to classify blood samples of patients suffering from blood cell disorders. Cells are detected using an R-CNN architecture. With the features extracted for each cell, a multiple instance learning method classifies patient samples into one out of four blood cell disorders. The attention mechanism provides a measure of the contribution of each cell to the overall classification and significantly improves the network's classification accuracy as well as its interpretability for the medical expert.
Scale-Space Autoencoders for Unsupervised Anomaly Segmentation in Brain MRI
Jun 23, 2020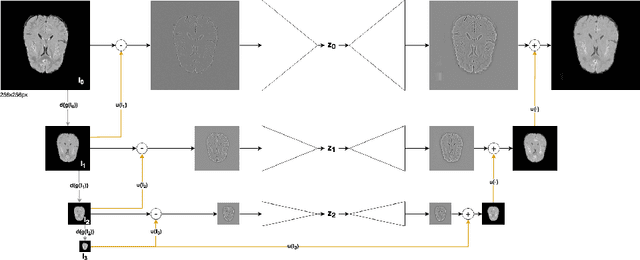
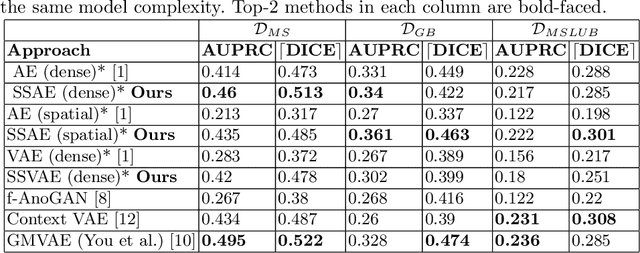
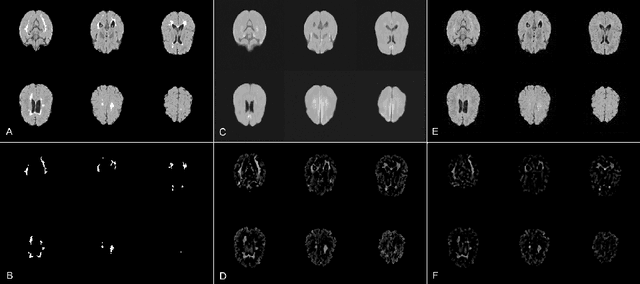
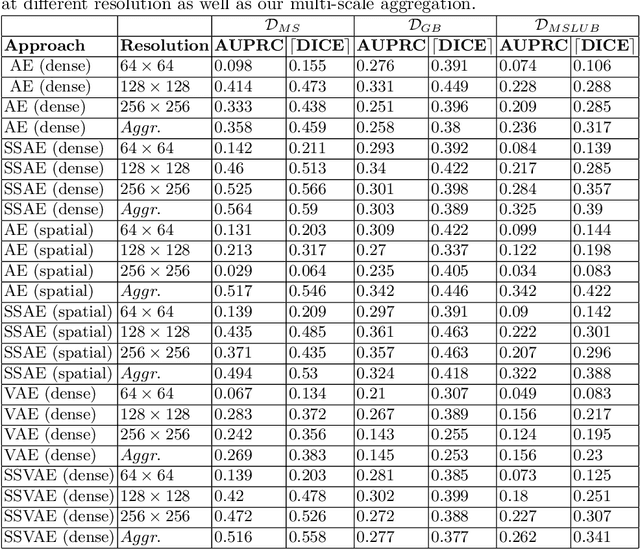
Abstract:Brain pathologies can vary greatly in size and shape, ranging from few pixels (i.e. MS lesions) to large, space-occupying tumors. Recently proposed Autoencoder-based methods for unsupervised anomaly segmentation in brain MRI have shown promising performance, but face difficulties in modeling distributions with high fidelity, which is crucial for accurate delineation of particularly small lesions. Here, similar to these previous works, we model the distribution of healthy brain MRI to localize pathologies from erroneous reconstructions. However, to achieve improved reconstruction fidelity at higher resolutions, we learn to compress and reconstruct different frequency bands of healthy brain MRI using the laplacian pyramid. In a range of experiments comparing our method to different State-of-the-Art approaches on three different brain MR datasets with MS lesions and tumors, we show improved anomaly segmentation performance and the general capability to obtain much more crisp reconstructions of input data at native resolution. The modeling of the laplacian pyramid further enables the delineation and aggregation of lesions at multiple scales, which allows to effectively cope with different pathologies and lesion sizes using a single model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge