Sandy Engelhardt
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Medical Diffusion -- Denoising Diffusion Probabilistic Models for 3D Medical Image Generation
Nov 07, 2022Abstract:Recent advances in computer vision have shown promising results in image generation. Diffusion probabilistic models in particular have generated realistic images from textual input, as demonstrated by DALL-E 2, Imagen and Stable Diffusion. However, their use in medicine, where image data typically comprises three-dimensional volumes, has not been systematically evaluated. Synthetic images may play a crucial role in privacy preserving artificial intelligence and can also be used to augment small datasets. Here we show that diffusion probabilistic models can synthesize high quality medical imaging data, which we show for Magnetic Resonance Images (MRI) and Computed Tomography (CT) images. We provide quantitative measurements of their performance through a reader study with two medical experts who rated the quality of the synthesized images in three categories: Realistic image appearance, anatomical correctness and consistency between slices. Furthermore, we demonstrate that synthetic images can be used in a self-supervised pre-training and improve the performance of breast segmentation models when data is scarce (dice score 0.91 vs. 0.95 without vs. with synthetic data).
Self-supervised motion descriptor for cardiac phase detection in 4D CMR based on discrete vector field estimations
Sep 18, 2022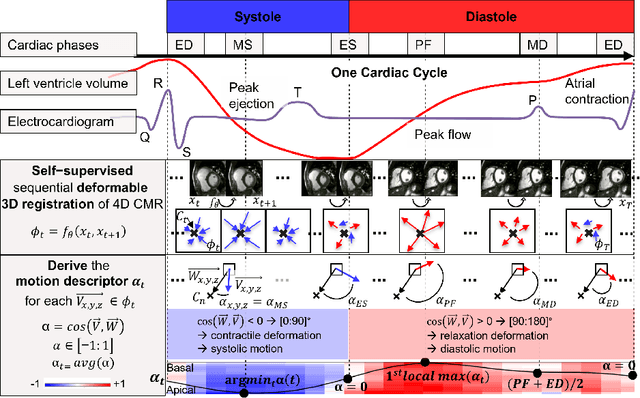
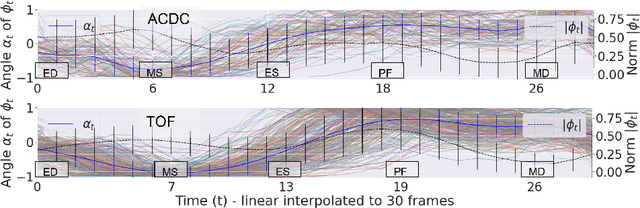
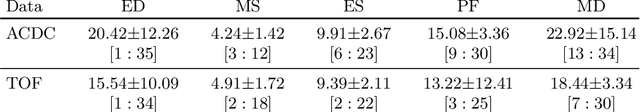
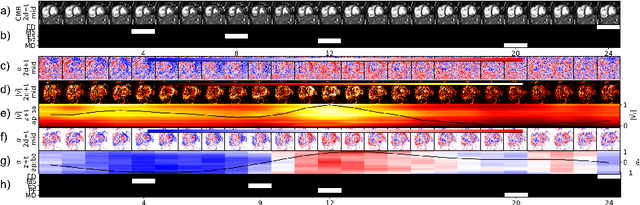
Abstract:Cardiac magnetic resonance (CMR) sequences visualise the cardiac function voxel-wise over time. Simultaneously, deep learning-based deformable image registration is able to estimate discrete vector fields which warp one time step of a CMR sequence to the following in a self-supervised manner. However, despite the rich source of information included in these 3D+t vector fields, a standardised interpretation is challenging and the clinical applications remain limited so far. In this work, we show how to efficiently use a deformable vector field to describe the underlying dynamic process of a cardiac cycle in form of a derived 1D motion descriptor. Additionally, based on the expected cardiovascular physiological properties of a contracting or relaxing ventricle, we define a set of rules that enables the identification of five cardiovascular phases including the end-systole (ES) and end-diastole (ED) without the usage of labels. We evaluate the plausibility of the motion descriptor on two challenging multi-disease, -center, -scanner short-axis CMR datasets. First, by reporting quantitative measures such as the periodic frame difference for the extracted phases. Second, by comparing qualitatively the general pattern when we temporally resample and align the motion descriptors of all instances across both datasets. The average periodic frame difference for the ED, ES key phases of our approach is $0.80\pm{0.85}$, $0.69\pm{0.79}$ which is slightly better than the inter-observer variability ($1.07\pm{0.86}$, $0.91\pm{1.6}$) and the supervised baseline method ($1.18\pm{1.91}$, $1.21\pm{1.78}$). Code and labels will be made available on our GitHub repository. https://github.com/Cardio-AI/cmr-phase-detection
Content-Aware Differential Privacy with Conditional Invertible Neural Networks
Jul 29, 2022
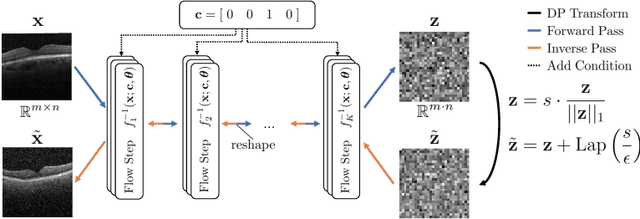
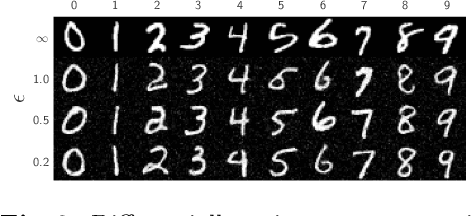
Abstract:Differential privacy (DP) has arisen as the gold standard in protecting an individual's privacy in datasets by adding calibrated noise to each data sample. While the application to categorical data is straightforward, its usability in the context of images has been limited. Contrary to categorical data the meaning of an image is inherent in the spatial correlation of neighboring pixels making the simple application of noise infeasible. Invertible Neural Networks (INN) have shown excellent generative performance while still providing the ability to quantify the exact likelihood. Their principle is based on transforming a complicated distribution into a simple one e.g. an image into a spherical Gaussian. We hypothesize that adding noise to the latent space of an INN can enable differentially private image modification. Manipulation of the latent space leads to a modified image while preserving important details. Further, by conditioning the INN on meta-data provided with the dataset we aim at leaving dimensions important for downstream tasks like classification untouched while altering other parts that potentially contain identifying information. We term our method content-aware differential privacy (CADP). We conduct experiments on publicly available benchmarking datasets as well as dedicated medical ones. In addition, we show the generalizability of our method to categorical data. The source code is publicly available at https://github.com/Cardio-AI/CADP.
mvHOTA: A multi-view higher order tracking accuracy metric to measure spatial and temporal associations in multi-point detection
Jun 19, 2022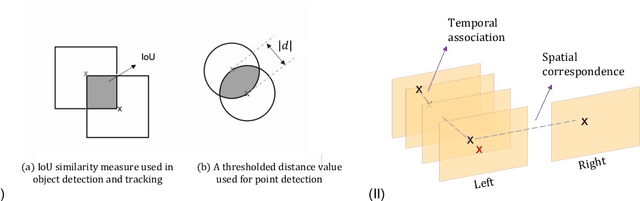
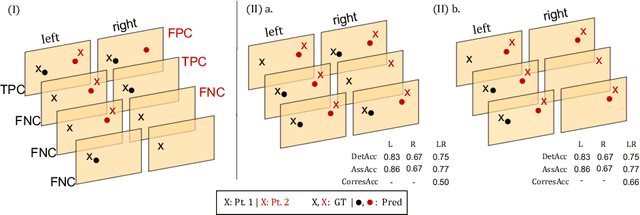
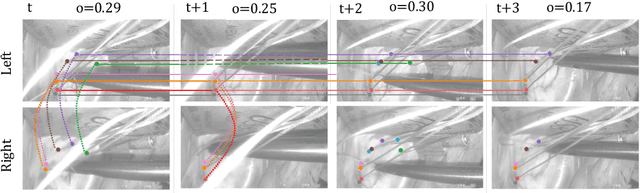
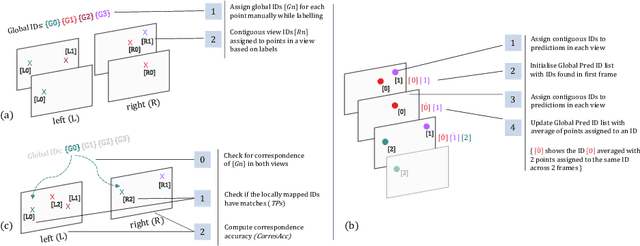
Abstract:Multi-object tracking (MOT) is a challenging task that involves detecting objects in the scene and tracking them across a sequence of frames. Evaluating this task is difficult due to temporal occlusions, and varying trajectories across a sequence of images. The main evaluation metric to benchmark MOT methods on datasets such as KITTI has recently become the higher order tracking accuracy (HOTA) metric, which is capable of providing a better description of the performance over metrics such as MOTA, DetA, and IDF1. Point detection and tracking is a closely related task, which could be regarded as a special case of object detection. However, there are differences in evaluating the detection task itself (point distances vs. bounding box overlap). When including the temporal dimension and multi-view scenarios, the evaluation task becomes even more complex. In this work, we propose a multi-view higher order tracking metric (mvHOTA) to determine the accuracy of multi-point (multi-instance and multi-class) detection, while taking into account temporal and spatial associations. mvHOTA can be interpreted as the geometric mean of the detection, association, and correspondence accuracies, thereby providing equal weighting to each of the factors. We demonstrate a use-case through a publicly available endoscopic point detection dataset from a previously organised medical challenge. Furthermore, we compare with other adjusted MOT metrics for this use-case, discuss the properties of mvHOTA, and show how the proposed correspondence accuracy and the Occlusion index facilitate analysis of methods with respect to handling of occlusions. The code will be made publicly available.
Posterior temperature optimized Bayesian models for inverse problems in medical imaging
Feb 02, 2022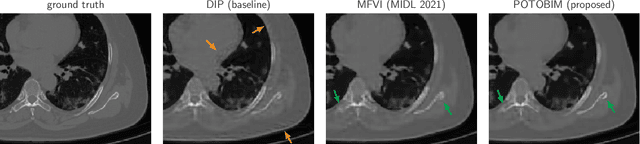
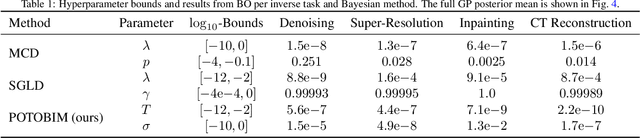
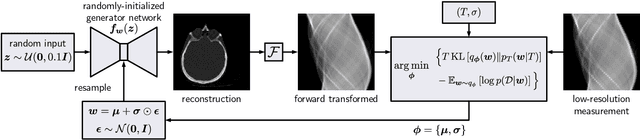
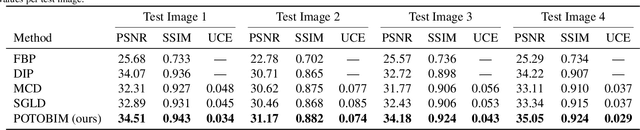
Abstract:We present Posterior Temperature Optimized Bayesian Inverse Models (POTOBIM), an unsupervised Bayesian approach to inverse problems in medical imaging using mean-field variational inference with a fully tempered posterior. Bayesian methods exhibit useful properties for approaching inverse tasks, such as tomographic reconstruction or image denoising. A suitable prior distribution introduces regularization, which is needed to solve the ill-posed problem and reduces overfitting the data. In practice, however, this often results in a suboptimal posterior temperature, and the full potential of the Bayesian approach is not being exploited. In POTOBIM, we optimize both the parameters of the prior distribution and the posterior temperature with respect to reconstruction accuracy using Bayesian optimization with Gaussian process regression. Our method is extensively evaluated on four different inverse tasks on a variety of modalities with images from public data sets and we demonstrate that an optimized posterior temperature outperforms both non-Bayesian and Bayesian approaches without temperature optimization. The use of an optimized prior distribution and posterior temperature leads to improved accuracy and uncertainty estimation and we show that it is sufficient to find these hyperparameters per task domain. Well-tempered posteriors yield calibrated uncertainty, which increases the reliability in the predictions. Our source code is publicly available at github.com/Cardio-AI/mfvi-dip-mia.
Comparison of Evaluation Metrics for Landmark Detection in CMR Images
Jan 28, 2022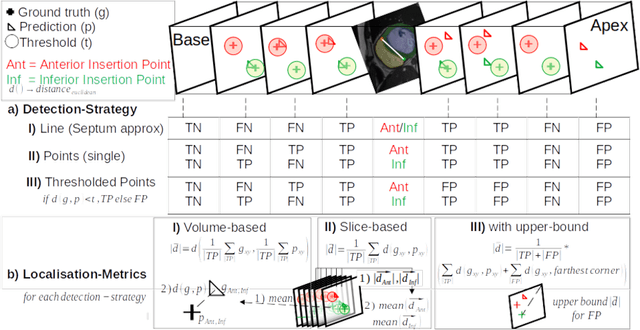
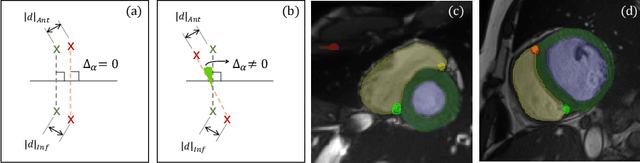
Abstract:Cardiac Magnetic Resonance (CMR) images are widely used for cardiac diagnosis and ventricular assessment. Extracting specific landmarks like the right ventricular insertion points is of importance for spatial alignment and 3D modeling. The automatic detection of such landmarks has been tackled by multiple groups using Deep Learning, but relatively little attention has been paid to the failure cases of evaluation metrics in this field. In this work, we extended the public ACDC dataset with additional labels of the right ventricular insertion points and compare different variants of a heatmap-based landmark detection pipeline. In this comparison, we demonstrate very likely pitfalls of apparently simple detection and localisation metrics which highlights the importance of a clear detection strategy and the definition of an upper limit for localisation-based metrics. Our preliminary results indicate that a combination of different metrics is necessary, as they yield different winners for method comparison. Additionally, they highlight the need of a comprehensive metric description and evaluation standardisation, especially for the error cases where no metrics could be computed or where no lower/upper boundary of a metric exists. Code and labels: https://github.com/Cardio-AI/rvip_landmark_detection
Comparison of Depth Estimation Setups from Stereo Endoscopy and Optical Tracking for Point Measurements
Jan 26, 2022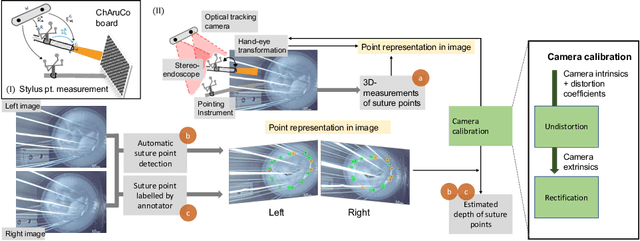

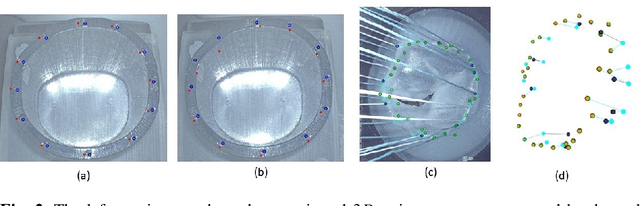
Abstract:To support minimally-invasive intraoperative mitral valve repair, quantitative measurements from the valve can be obtained using an infra-red tracked stylus. It is desirable to view such manually measured points together with the endoscopic image for further assistance. Therefore, hand-eye calibration is required that links both coordinate systems and is a prerequisite to project the points onto the image plane. A complementary approach to this is to use a vision-based endoscopic stereo-setup to detect and triangulate points of interest, to obtain the 3D coordinates. In this paper, we aim to compare both approaches on a rigid phantom and two patient-individual silicone replica which resemble the intraoperative scenario. The preliminary results indicate that 3D landmark estimation, either labeled manually or through partly automated detection with a deep learning approach, provides more accurate triangulated depth measurements when performed with a tailored image-based method than with stylus measurements.
Point detection through multi-instance deep heatmap regression for sutures in endoscopy
Nov 16, 2021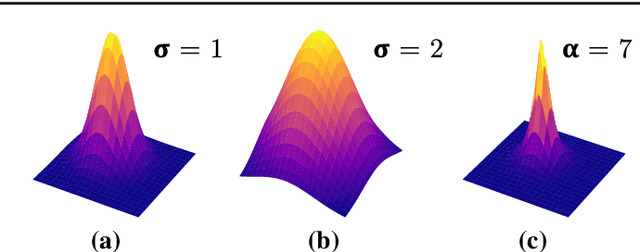
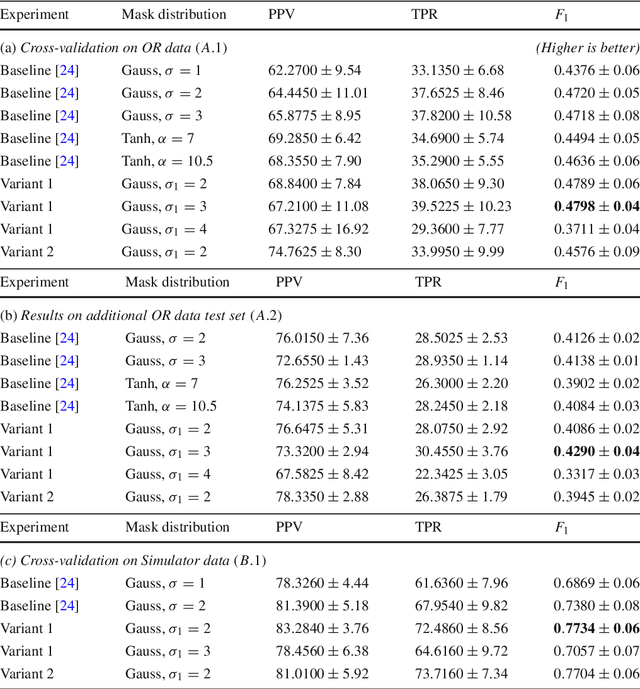
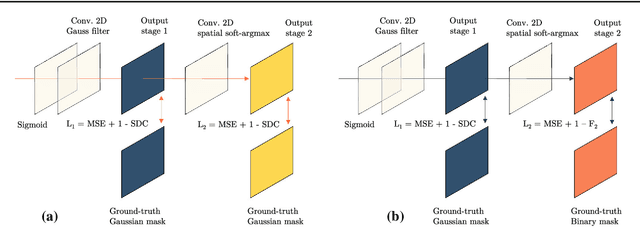
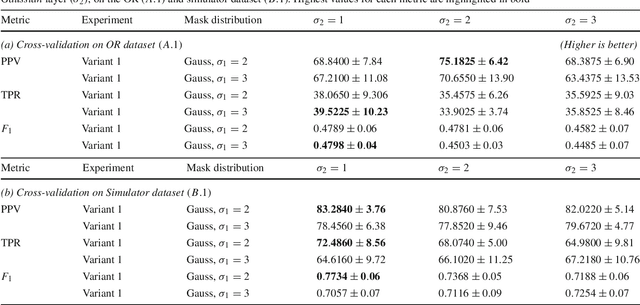
Abstract:Purpose: Mitral valve repair is a complex minimally invasive surgery of the heart valve. In this context, suture detection from endoscopic images is a highly relevant task that provides quantitative information to analyse suturing patterns, assess prosthetic configurations and produce augmented reality visualisations. Facial or anatomical landmark detection tasks typically contain a fixed number of landmarks, and use regression or fixed heatmap-based approaches to localize the landmarks. However in endoscopy, there are a varying number of sutures in every image, and the sutures may occur at any location in the annulus, as they are not semantically unique. Method: In this work, we formulate the suture detection task as a multi-instance deep heatmap regression problem, to identify entry and exit points of sutures. We extend our previous work, and introduce the novel use of a 2D Gaussian layer followed by a differentiable 2D spatial Soft-Argmax layer to function as a local non-maximum suppression. Results: We present extensive experiments with multiple heatmap distribution functions and two variants of the proposed model. In the intra-operative domain, Variant 1 showed a mean F1 of +0.0422 over the baseline. Similarly, in the simulator domain, Variant 1 showed a mean F1 of +0.0865 over the baseline. Conclusion: The proposed model shows an improvement over the baseline in the intra-operative and the simulator domains. The data is made publicly available within the scope of the MICCAI AdaptOR2021 Challenge https://adaptor2021.github.io/, and the code at https://github.com/Cardio-AI/suture-detection-pytorch/. DOI:10.1007/s11548-021-02523-w. The link to the open access article can be found here: https://link.springer.com/article/10.1007%2Fs11548-021-02523-w
* Accepted to International Journal of Computer Assisted Radiology and Surgery, 15 pages, 5 figures
Mutually improved endoscopic image synthesis and landmark detection in unpaired image-to-image translation
Jul 14, 2021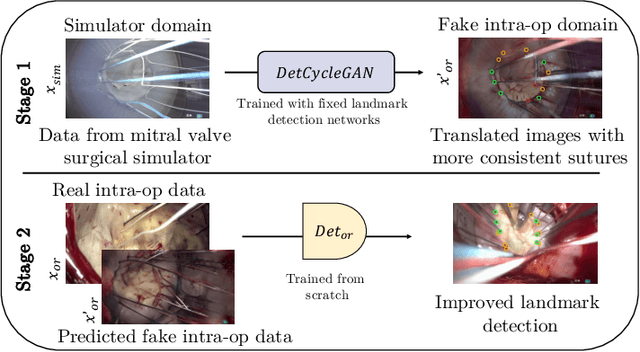
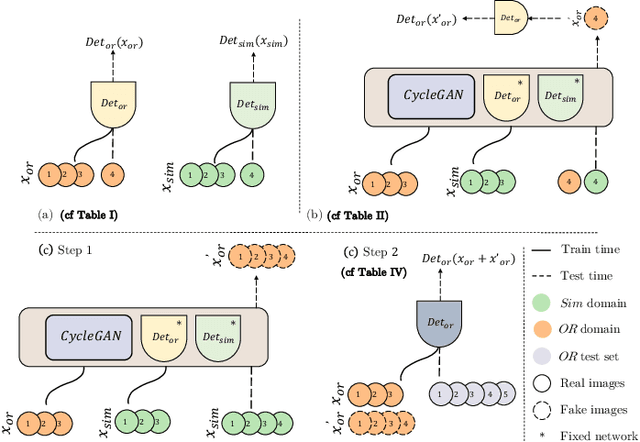
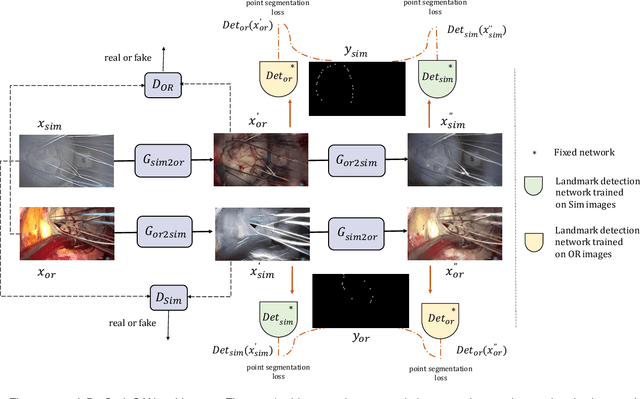
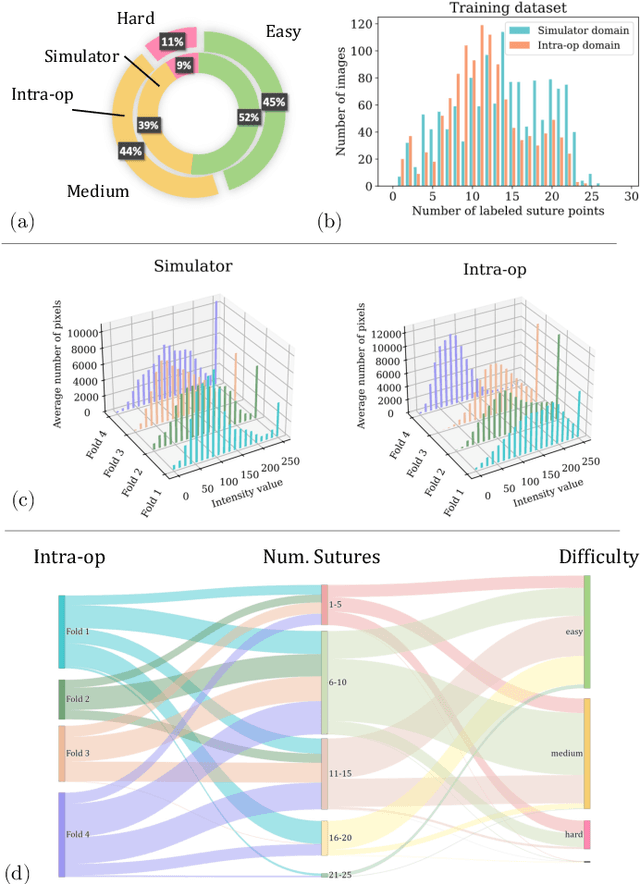
Abstract:The CycleGAN framework allows for unsupervised image-to-image translation of unpaired data. In a scenario of surgical training on a physical surgical simulator, this method can be used to transform endoscopic images of phantoms into images which more closely resemble the intra-operative appearance of the same surgical target structure. This can be viewed as a novel augmented reality approach, which we coined Hyperrealism in previous work. In this use case, it is of paramount importance to display objects like needles, sutures or instruments consistent in both domains while altering the style to a more tissue-like appearance. Segmentation of these objects would allow for a direct transfer, however, contouring of these, partly tiny and thin foreground objects is cumbersome and perhaps inaccurate. Instead, we propose to use landmark detection on the points when sutures pass into the tissue. This objective is directly incorporated into a CycleGAN framework by treating the performance of pre-trained detector models as an additional optimization goal. We show that a task defined on these sparse landmark labels improves consistency of synthesis by the generator network in both domains. Comparing a baseline CycleGAN architecture to our proposed extension (DetCycleGAN), mean precision (PPV) improved by +61.32, mean sensitivity (TPR) by +37.91, and mean F1 score by +0.4743. Furthermore, it could be shown that by dataset fusion, generated intra-operative images can be leveraged as additional training data for the detection network itself. The data is released within the scope of the AdaptOR MICCAI Challenge 2021 at https://adaptor2021.github.io/, and code at https://github.com/Cardio-AI/detcyclegan_pytorch.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge