Robyn Ball
A Multi-agent Large Language Model Framework to Automatically Assess Performance of a Clinical AI Triage Tool
Oct 30, 2025Abstract:Purpose: The purpose of this study was to determine if an ensemble of multiple LLM agents could be used collectively to provide a more reliable assessment of a pixel-based AI triage tool than a single LLM. Methods: 29,766 non-contrast CT head exams from fourteen hospitals were processed by a commercial intracranial hemorrhage (ICH) AI detection tool. Radiology reports were analyzed by an ensemble of eight open-source LLM models and a HIPAA compliant internal version of GPT-4o using a single multi-shot prompt that assessed for presence of ICH. 1,726 examples were manually reviewed. Performance characteristics of the eight open-source models and consensus were compared to GPT-4o. Three ideal consensus LLM ensembles were tested for rating the performance of the triage tool. Results: The cohort consisted of 29,766 head CTs exam-report pairs. The highest AUC performance was achieved with llama3.3:70b and GPT-4o (AUC= 0.78). The average precision was highest for Llama3.3:70b and GPT-4o (AP=0.75 & 0.76). Llama3.3:70b had the highest F1 score (0.81) and recall (0.85), greater precision (0.78), specificity (0.72), and MCC (0.57). Using MCC (95% CI) the ideal combination of LLMs were: Full-9 Ensemble 0.571 (0.552-0.591), Top-3 Ensemble 0.558 (0.537-0.579), Consensus 0.556 (0.539-0.574), and GPT4o 0.522 (0.500-0.543). No statistically significant differences were observed between Top-3, Full-9, and Consensus (p > 0.05). Conclusion: An ensemble of medium to large sized open-source LLMs provides a more consistent and reliable method to derive a ground truth retrospective evaluation of a clinical AI triage tool over a single LLM alone.
CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison
Jan 21, 2019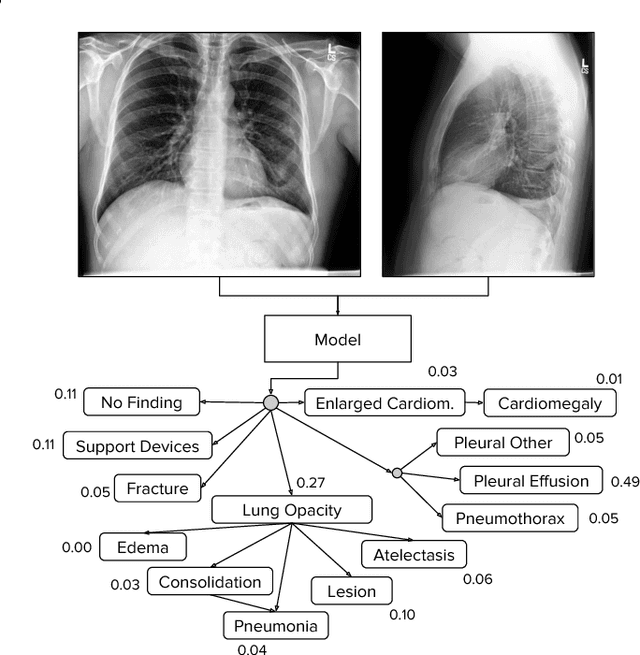
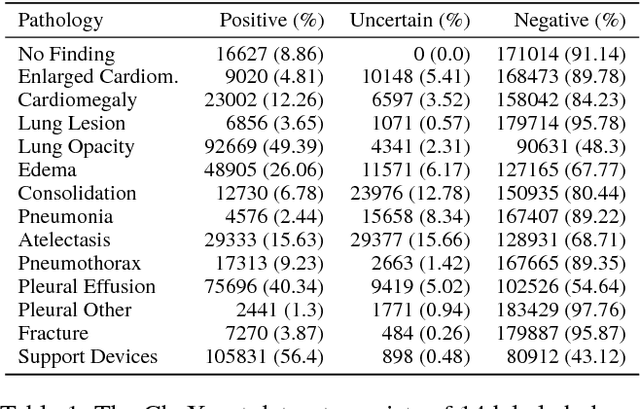
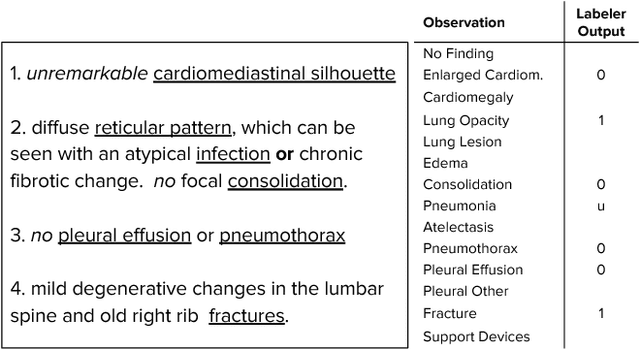
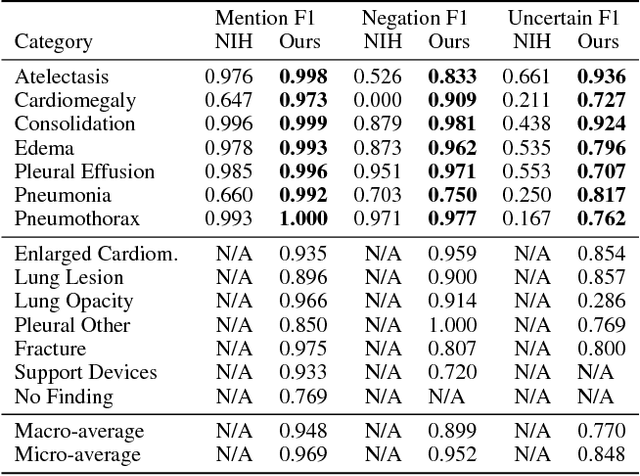
Abstract:Large, labeled datasets have driven deep learning methods to achieve expert-level performance on a variety of medical imaging tasks. We present CheXpert, a large dataset that contains 224,316 chest radiographs of 65,240 patients. We design a labeler to automatically detect the presence of 14 observations in radiology reports, capturing uncertainties inherent in radiograph interpretation. We investigate different approaches to using the uncertainty labels for training convolutional neural networks that output the probability of these observations given the available frontal and lateral radiographs. On a validation set of 200 chest radiographic studies which were manually annotated by 3 board-certified radiologists, we find that different uncertainty approaches are useful for different pathologies. We then evaluate our best model on a test set composed of 500 chest radiographic studies annotated by a consensus of 5 board-certified radiologists, and compare the performance of our model to that of 3 additional radiologists in the detection of 5 selected pathologies. On Cardiomegaly, Edema, and Pleural Effusion, the model ROC and PR curves lie above all 3 radiologist operating points. We release the dataset to the public as a standard benchmark to evaluate performance of chest radiograph interpretation models. The dataset is freely available at https://stanfordmlgroup.github.io/competitions/chexpert .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge