Po-Chih Kuo
Incorporating Eye-Tracking Signals Into Multimodal Deep Visual Models For Predicting User Aesthetic Experience In Residential Interiors
Jan 23, 2026Abstract:Understanding how people perceive and evaluate interior spaces is essential for designing environments that promote well-being. However, predicting aesthetic experiences remains difficult due to the subjective nature of perception and the complexity of visual responses. This study introduces a dual-branch CNN-LSTM framework that fuses visual features with eye-tracking signals to predict aesthetic evaluations of residential interiors. We collected a dataset of 224 interior design videos paired with synchronized gaze data from 28 participants who rated 15 aesthetic dimensions. The proposed model attains 72.2% accuracy on objective dimensions (e.g., light) and 66.8% on subjective dimensions (e.g., relaxation), outperforming state-of-the-art video baselines and showing clear gains on subjective evaluation tasks. Notably, models trained with eye-tracking retain comparable performance when deployed with visual input alone. Ablation experiments further reveal that pupil responses contribute most to objective assessments, while the combination of gaze and visual cues enhances subjective evaluations. These findings highlight the value of incorporating eye-tracking as privileged information during training, enabling more practical tools for aesthetic assessment in interior design.
Algorithms Trained on Normal Chest X-rays Can Predict Health Insurance Types
Nov 17, 2025Abstract:Artificial intelligence is revealing what medicine never intended to encode. Deep vision models, trained on chest X-rays, can now detect not only disease but also invisible traces of social inequality. In this study, we show that state-of-the-art architectures (DenseNet121, SwinV2-B, MedMamba) can predict a patient's health insurance type, a strong proxy for socioeconomic status, from normal chest X-rays with significant accuracy (AUC around 0.67 on MIMIC-CXR-JPG, 0.68 on CheXpert). The signal persists even when age, race, and sex are controlled for, and remains detectable when the model is trained exclusively on a single racial group. Patch-based occlusion reveals that the signal is diffuse rather than localized, embedded in the upper and mid-thoracic regions. This suggests that deep networks may be internalizing subtle traces of clinical environments, equipment differences, or care pathways; learning socioeconomic segregation itself. These findings challenge the assumption that medical images are neutral biological data. By uncovering how models perceive and exploit these hidden social signatures, this work reframes fairness in medical AI: the goal is no longer only to balance datasets or adjust thresholds, but to interrogate and disentangle the social fingerprints embedded in clinical data itself.
Uncovering Overconfident Failures in CXR Models via Augmentation-Sensitivity Risk Scoring
Oct 02, 2025Abstract:Deep learning models achieve strong performance in chest radiograph (CXR) interpretation, yet fairness and reliability concerns persist. Models often show uneven accuracy across patient subgroups, leading to hidden failures not reflected in aggregate metrics. Existing error detection approaches -- based on confidence calibration or out-of-distribution (OOD) detection -- struggle with subtle within-distribution errors, while image- and representation-level consistency-based methods remain underexplored in medical imaging. We propose an augmentation-sensitivity risk scoring (ASRS) framework to identify error-prone CXR cases. ASRS applies clinically plausible rotations ($\pm 15^\circ$/$\pm 30^\circ$) and measures embedding shifts with the RAD-DINO encoder. Sensitivity scores stratify samples into stability quartiles, where highly sensitive cases show substantially lower recall ($-0.2$ to $-0.3$) despite high AUROC and confidence. ASRS provides a label-free means for selective prediction and clinician review, improving fairness and safety in medical AI.
LTCXNet: Advancing Chest X-Ray Analysis with Solutions for Long-Tailed Multi-Label Classification and Fairness Challenges
Nov 16, 2024Abstract:Chest X-rays (CXRs) often display various diseases with disparate class frequencies, leading to a long-tailed, multi-label data distribution. In response to this challenge, we explore the Pruned MIMIC-CXR-LT dataset, a curated collection derived from the MIMIC-CXR dataset, specifically designed to represent a long-tailed and multi-label data scenario. We introduce LTCXNet, a novel framework that integrates the ConvNeXt model, ML-Decoder, and strategic data augmentation, further enhanced by an ensemble approach. We demonstrate that LTCXNet improves the performance of CXR interpretation across all classes, especially enhancing detection in rarer classes like `Pneumoperitoneum' and `Pneumomediastinum' by 79\% and 48\%, respectively. Beyond performance metrics, our research extends into evaluating fairness, highlighting that some methods, while improving model accuracy, could inadvertently affect fairness across different demographic groups negatively. This work contributes to advancing the understanding and management of long-tailed, multi-label data distributions in medical imaging, paving the way for more equitable and effective diagnostic tools.
DengueNet: Dengue Prediction using Spatiotemporal Satellite Imagery for Resource-Limited Countries
Jan 23, 2024



Abstract:Dengue fever presents a substantial challenge in developing countries where sanitation infrastructure is inadequate. The absence of comprehensive healthcare systems exacerbates the severity of dengue infections, potentially leading to life-threatening circumstances. Rapid response to dengue outbreaks is also challenging due to limited information exchange and integration. While timely dengue outbreak forecasts have the potential to prevent such outbreaks, the majority of dengue prediction studies have predominantly relied on data that impose significant burdens on individual countries for collection. In this study, our aim is to improve health equity in resource-constrained countries by exploring the effectiveness of high-resolution satellite imagery as a nontraditional and readily accessible data source. By leveraging the wealth of publicly available and easily obtainable satellite imagery, we present a scalable satellite extraction framework based on Sentinel Hub, a cloud-based computing platform. Furthermore, we introduce DengueNet, an innovative architecture that combines Vision Transformer, Radiomics, and Long Short-term Memory to extract and integrate spatiotemporal features from satellite images. This enables dengue predictions on an epi-week basis. To evaluate the effectiveness of our proposed method, we conducted experiments on five municipalities in Colombia. We utilized a dataset comprising 780 high-resolution Sentinel-2 satellite images for training and evaluation. The performance of DengueNet was assessed using the mean absolute error (MAE) metric. Across the five municipalities, DengueNet achieved an average MAE of 43.92. Our findings strongly support the efficacy of satellite imagery as a valuable resource for dengue prediction, particularly in informing public health policies within countries where manually collected data is scarce and dengue virus prevalence is severe.
Dementia Assessment Using Mandarin Speech with an Attention-based Speech Recognition Encoder
Oct 06, 2023Abstract:Dementia diagnosis requires a series of different testing methods, which is complex and time-consuming. Early detection of dementia is crucial as it can prevent further deterioration of the condition. This paper utilizes a speech recognition model to construct a dementia assessment system tailored for Mandarin speakers during the picture description task. By training an attention-based speech recognition model on voice data closely resembling real-world scenarios, we have significantly enhanced the model's recognition capabilities. Subsequently, we extracted the encoder from the speech recognition model and added a linear layer for dementia assessment. We collected Mandarin speech data from 99 subjects and acquired their clinical assessments from a local hospital. We achieved an accuracy of 92.04% in Alzheimer's disease detection and a mean absolute error of 9% in clinical dementia rating score prediction.
Early Diagnosis of Chronic Obstructive Pulmonary Disease from Chest X-Rays using Transfer Learning and Fusion Strategies
Nov 13, 2022Abstract:Chronic obstructive pulmonary disease (COPD) is one of the most common chronic illnesses in the world and the third leading cause of mortality worldwide. It is often underdiagnosed or not diagnosed until later in the disease course. Spirometry tests are the gold standard for diagnosing COPD but can be difficult to obtain, especially in resource-poor countries. Chest X-rays (CXRs), however, are readily available and may serve as a screening tool to identify patients with COPD who should undergo further testing. Currently, no research applies deep learning (DL) algorithms that use large multi-site and multi-modal data to detect COPD patients and evaluate fairness across demographic groups. We use three CXR datasets in our study, CheXpert to pre-train models, MIMIC-CXR to develop, and Emory-CXR to validate our models. The CXRs from patients in the early stage of COPD and not on mechanical ventilation are selected for model training and validation. We visualize the Grad-CAM heatmaps of the true positive cases on the base model for both MIMIC-CXR and Emory-CXR test datasets. We further propose two fusion schemes, (1) model-level fusion, including bagging and stacking methods using MIMIC-CXR, and (2) data-level fusion, including multi-site data using MIMIC-CXR and Emory-CXR, and multi-modal using MIMIC-CXRs and MIMIC-IV EHR, to improve the overall model performance. Fairness analysis is performed to evaluate if the fusion schemes have a discrepancy in the performance among different demographic groups. The results demonstrate that DL models can detect COPD using CXRs, which can facilitate early screening, especially in low-resource regions where CXRs are more accessible than spirometry. The multi-site data fusion scheme could improve the model generalizability on the Emory-CXR test data. Further studies on using CXR or other modalities to predict COPD ought to be in future work.
Developing and validating multi-modal models for mortality prediction in COVID-19 patients: a multi-center retrospective study
Sep 01, 2021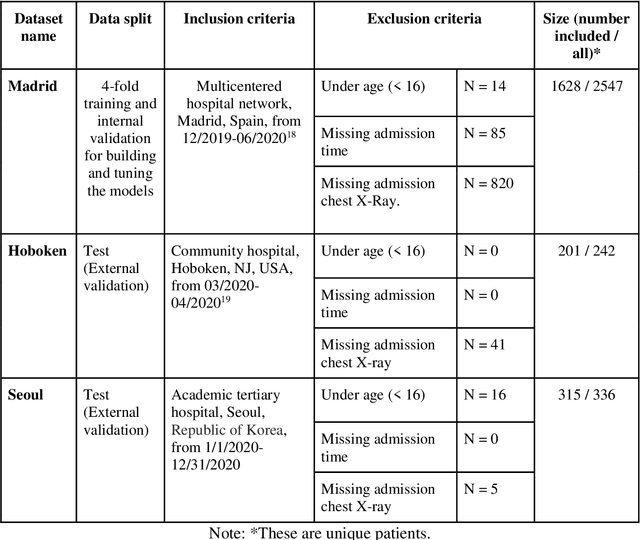
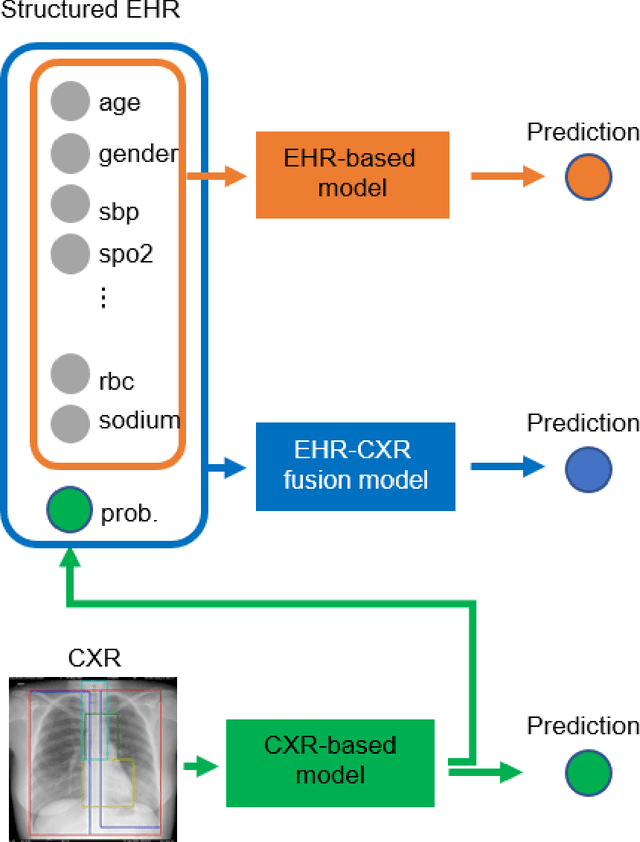
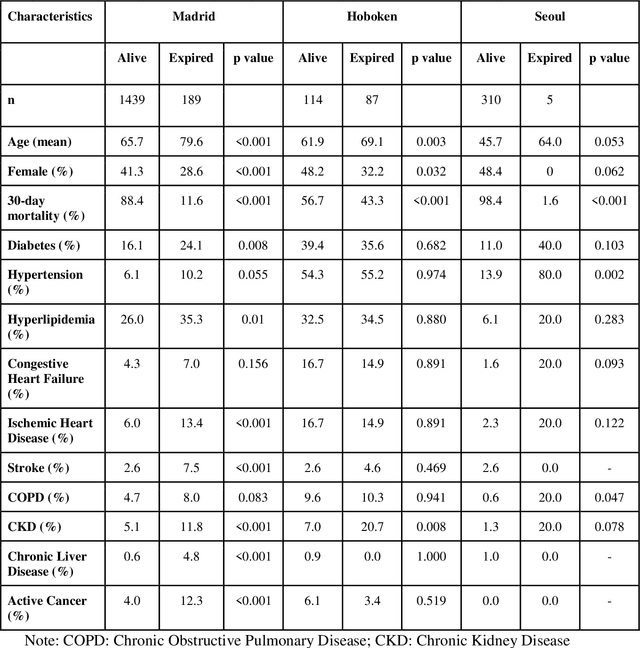
Abstract:The unprecedented global crisis brought about by the COVID-19 pandemic has sparked numerous efforts to create predictive models for the detection and prognostication of SARS-CoV-2 infections with the goal of helping health systems allocate resources. Machine learning models, in particular, hold promise for their ability to leverage patient clinical information and medical images for prediction. However, most of the published COVID-19 prediction models thus far have little clinical utility due to methodological flaws and lack of appropriate validation. In this paper, we describe our methodology to develop and validate multi-modal models for COVID-19 mortality prediction using multi-center patient data. The models for COVID-19 mortality prediction were developed using retrospective data from Madrid, Spain (N=2547) and were externally validated in patient cohorts from a community hospital in New Jersey, USA (N=242) and an academic center in Seoul, Republic of Korea (N=336). The models we developed performed differently across various clinical settings, underscoring the need for a guided strategy when employing machine learning for clinical decision-making. We demonstrated that using features from both the structured electronic health records and chest X-ray imaging data resulted in better 30-day-mortality prediction performance across all three datasets (areas under the receiver operating characteristic curves: 0.85 (95% confidence interval: 0.83-0.87), 0.76 (0.70-0.82), and 0.95 (0.92-0.98)). We discuss the rationale for the decisions made at every step in developing the models and have made our code available to the research community. We employed the best machine learning practices for clinical model development. Our goal is to create a toolkit that would assist investigators and organizations in building multi-modal models for prediction, classification and/or optimization.
Reading Race: AI Recognises Patient's Racial Identity In Medical Images
Jul 21, 2021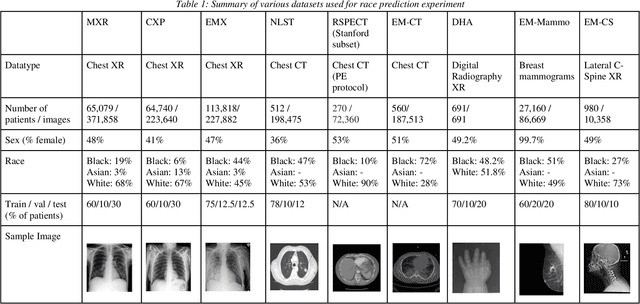
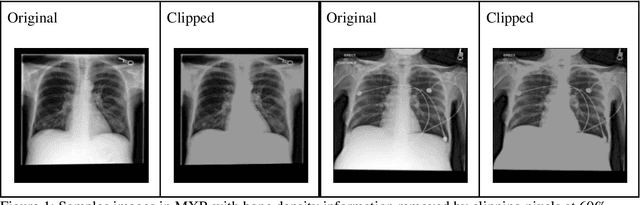
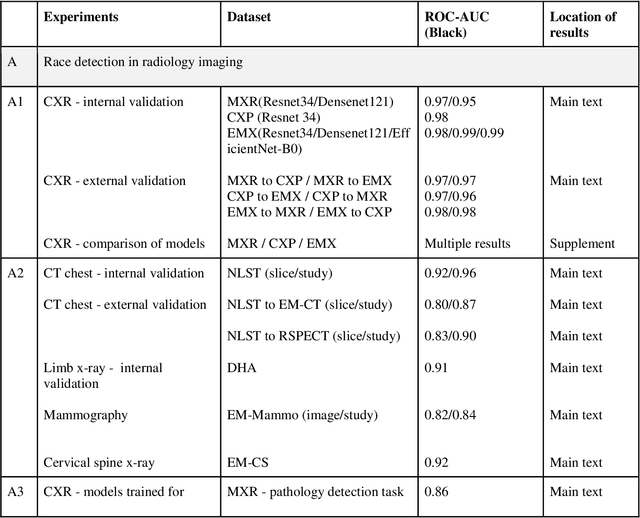
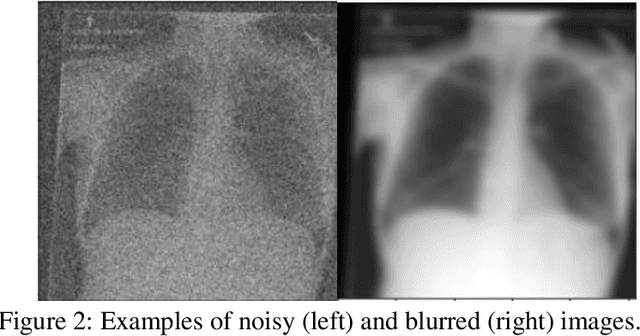
Abstract:Background: In medical imaging, prior studies have demonstrated disparate AI performance by race, yet there is no known correlation for race on medical imaging that would be obvious to the human expert interpreting the images. Methods: Using private and public datasets we evaluate: A) performance quantification of deep learning models to detect race from medical images, including the ability of these models to generalize to external environments and across multiple imaging modalities, B) assessment of possible confounding anatomic and phenotype population features, such as disease distribution and body habitus as predictors of race, and C) investigation into the underlying mechanism by which AI models can recognize race. Findings: Standard deep learning models can be trained to predict race from medical images with high performance across multiple imaging modalities. Our findings hold under external validation conditions, as well as when models are optimized to perform clinically motivated tasks. We demonstrate this detection is not due to trivial proxies or imaging-related surrogate covariates for race, such as underlying disease distribution. Finally, we show that performance persists over all anatomical regions and frequency spectrum of the images suggesting that mitigation efforts will be challenging and demand further study. Interpretation: We emphasize that model ability to predict self-reported race is itself not the issue of importance. However, our findings that AI can trivially predict self-reported race -- even from corrupted, cropped, and noised medical images -- in a setting where clinical experts cannot, creates an enormous risk for all model deployments in medical imaging: if an AI model secretly used its knowledge of self-reported race to misclassify all Black patients, radiologists would not be able to tell using the same data the model has access to.
Interpretable Machine Learning Model for Early Prediction of Mortality in Elderly Patients with Multiple Organ Dysfunction Syndrome (MODS): a Multicenter Retrospective Study and Cross Validation
Jan 28, 2020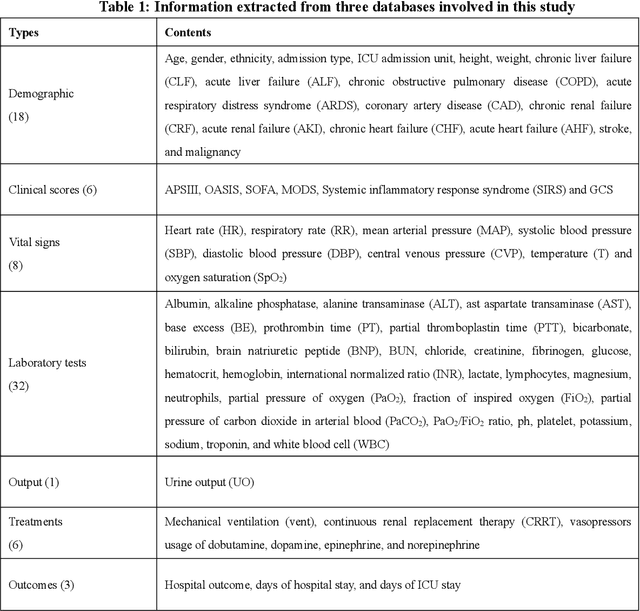
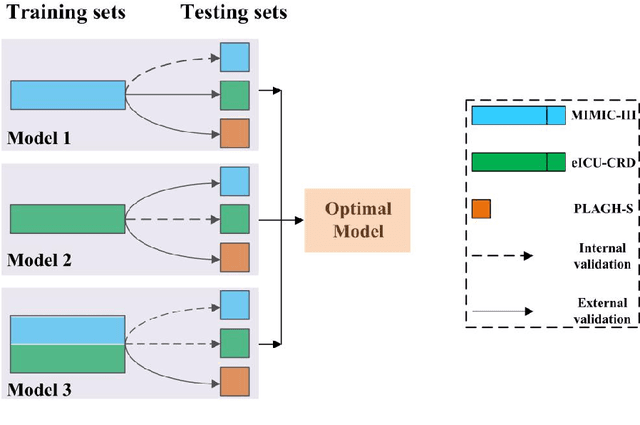
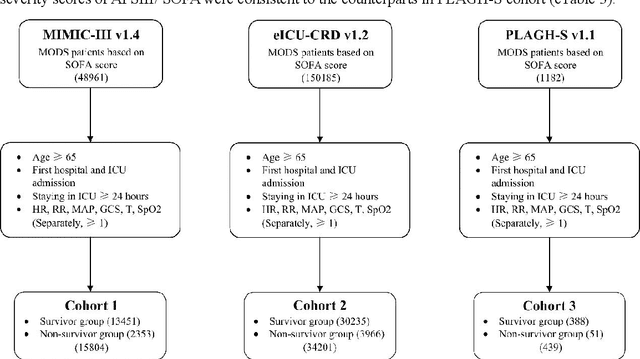
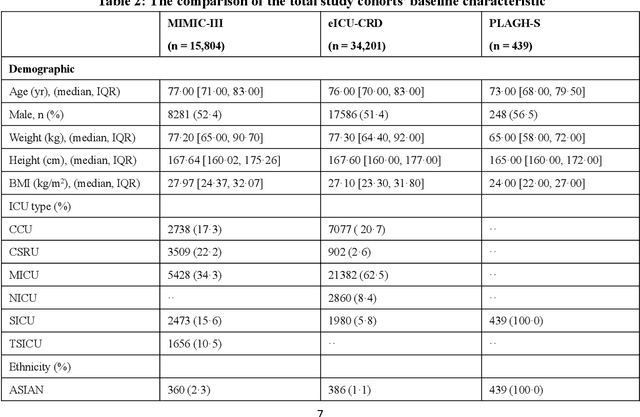
Abstract:Background: Elderly patients with MODS have high risk of death and poor prognosis. The performance of current scoring systems assessing the severity of MODS and its mortality remains unsatisfactory. This study aims to develop an interpretable and generalizable model for early mortality prediction in elderly patients with MODS. Methods: The MIMIC-III, eICU-CRD and PLAGH-S databases were employed for model generation and evaluation. We used the eXtreme Gradient Boosting model with the SHapley Additive exPlanations method to conduct early and interpretable predictions of patients' hospital outcome. Three types of data source combinations and five typical evaluation indexes were adopted to develop a generalizable model. Findings: The interpretable model, with optimal performance developed by using MIMIC-III and eICU-CRD datasets, was separately validated in MIMIC-III, eICU-CRD and PLAGH-S datasets (no overlapping with training set). The performances of the model in predicting hospital mortality as validated by the three datasets were: AUC of 0.858, sensitivity of 0.834 and specificity of 0.705; AUC of 0.849, sensitivity of 0.763 and specificity of 0.784; and AUC of 0.838, sensitivity of 0.882 and specificity of 0.691, respectively. Comparisons of AUC between this model and baseline models with MIMIC-III dataset validation showed superior performances of this model; In addition, comparisons in AUC between this model and commonly used clinical scores showed significantly better performance of this model. Interpretation: The interpretable machine learning model developed in this study using fused datasets with large sample sizes was robust and generalizable. This model outperformed the baseline models and several clinical scores for early prediction of mortality in elderly ICU patients. The interpretative nature of this model provided clinicians with the ranking of mortality risk features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge