Pekka Ruusuvuori
Institute of Biomedicine, University of Turku, Turku, Finland and InFLAMES Research Flagship, University of Turku, Turku, Finland and Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland
Staining normalization in histopathology: Method benchmarking using multicenter dataset
Jun 23, 2025Abstract:Hematoxylin and Eosin (H&E) has been the gold standard in tissue analysis for decades, however, tissue specimens stained in different laboratories vary, often significantly, in appearance. This variation poses a challenge for both pathologists' and AI-based downstream analysis. Minimizing stain variation computationally is an active area of research. To further investigate this problem, we collected a unique multi-center tissue image dataset, wherein tissue samples from colon, kidney, and skin tissue blocks were distributed to 66 different labs for routine H&E staining. To isolate staining variation, other factors affecting the tissue appearance were kept constant. Further, we used this tissue image dataset to compare the performance of eight different stain normalization methods, including four traditional methods, namely, histogram matching, Macenko, Vahadane, and Reinhard normalization, and two deep learning-based methods namely CycleGAN and Pixp2pix, both with two variants each. We used both quantitative and qualitative evaluation to assess the performance of these methods. The dataset's inter-laboratory staining variation could also guide strategies to improve model generalizability through varied training data
Foundation Models -- A Panacea for Artificial Intelligence in Pathology?
Feb 28, 2025



Abstract:The role of artificial intelligence (AI) in pathology has evolved from aiding diagnostics to uncovering predictive morphological patterns in whole slide images (WSIs). Recently, foundation models (FMs) leveraging self-supervised pre-training have been widely advocated as a universal solution for diverse downstream tasks. However, open questions remain about their clinical applicability and generalization advantages over end-to-end learning using task-specific (TS) models. Here, we focused on AI with clinical-grade performance for prostate cancer diagnosis and Gleason grading. We present the largest validation of AI for this task, using over 100,000 core needle biopsies from 7,342 patients across 15 sites in 11 countries. We compared two FMs with a fully end-to-end TS model in a multiple instance learning framework. Our findings challenge assumptions that FMs universally outperform TS models. While FMs demonstrated utility in data-scarce scenarios, their performance converged with - and was in some cases surpassed by - TS models when sufficient labeled training data were available. Notably, extensive task-specific training markedly reduced clinically significant misgrading, misdiagnosis of challenging morphologies, and variability across different WSI scanners. Additionally, FMs used up to 35 times more energy than the TS model, raising concerns about their sustainability. Our results underscore that while FMs offer clear advantages for rapid prototyping and research, their role as a universal solution for clinically applicable medical AI remains uncertain. For high-stakes clinical applications, rigorous validation and consideration of task-specific training remain critically important. We advocate for integrating the strengths of FMs and end-to-end learning to achieve robust and resource-efficient AI pathology solutions fit for clinical use.
Improving Performance in Colorectal Cancer Histology Decomposition using Deep and Ensemble Machine Learning
Oct 25, 2023



Abstract:In routine colorectal cancer management, histologic samples stained with hematoxylin and eosin are commonly used. Nonetheless, their potential for defining objective biomarkers for patient stratification and treatment selection is still being explored. The current gold standard relies on expensive and time-consuming genetic tests. However, recent research highlights the potential of convolutional neural networks (CNNs) in facilitating the extraction of clinically relevant biomarkers from these readily available images. These CNN-based biomarkers can predict patient outcomes comparably to golden standards, with the added advantages of speed, automation, and minimal cost. The predictive potential of CNN-based biomarkers fundamentally relies on the ability of convolutional neural networks (CNNs) to classify diverse tissue types from whole slide microscope images accurately. Consequently, enhancing the accuracy of tissue class decomposition is critical to amplifying the prognostic potential of imaging-based biomarkers. This study introduces a hybrid Deep and ensemble machine learning model that surpassed all preceding solutions for this classification task. Our model achieved 96.74% accuracy on the external test set and 99.89% on the internal test set. Recognizing the potential of these models in advancing the task, we have made them publicly available for further research and development.
Physical Color Calibration of Digital Pathology Scanners for Robust Artificial Intelligence Assisted Cancer Diagnosis
Jul 07, 2023



Abstract:The potential of artificial intelligence (AI) in digital pathology is limited by technical inconsistencies in the production of whole slide images (WSIs), leading to degraded AI performance and posing a challenge for widespread clinical application as fine-tuning algorithms for each new site is impractical. Changes in the imaging workflow can also lead to compromised diagnoses and patient safety risks. We evaluated whether physical color calibration of scanners can standardize WSI appearance and enable robust AI performance. We employed a color calibration slide in four different laboratories and evaluated its impact on the performance of an AI system for prostate cancer diagnosis on 1,161 WSIs. Color standardization resulted in consistently improved AI model calibration and significant improvements in Gleason grading performance. The study demonstrates that physical color calibration provides a potential solution to the variation introduced by different scanners, making AI-based cancer diagnostics more reliable and applicable in clinical settings.
The ACROBAT 2022 Challenge: Automatic Registration Of Breast Cancer Tissue
May 29, 2023Abstract:The alignment of tissue between histopathological whole-slide-images (WSI) is crucial for research and clinical applications. Advances in computing, deep learning, and availability of large WSI datasets have revolutionised WSI analysis. Therefore, the current state-of-the-art in WSI registration is unclear. To address this, we conducted the ACROBAT challenge, based on the largest WSI registration dataset to date, including 4,212 WSIs from 1,152 breast cancer patients. The challenge objective was to align WSIs of tissue that was stained with routine diagnostic immunohistochemistry to its H&E-stained counterpart. We compare the performance of eight WSI registration algorithms, including an investigation of the impact of different WSI properties and clinical covariates. We find that conceptually distinct WSI registration methods can lead to highly accurate registration performances and identify covariates that impact performances across methods. These results establish the current state-of-the-art in WSI registration and guide researchers in selecting and developing methods.
Domain-specific transfer learning in the automated scoring of tumor-stroma ratio from histopathological images of colorectal cancer
Dec 30, 2022Abstract:Tumor-stroma ratio (TSR) is a prognostic factor for many types of solid tumors. In this study, we propose a method for automated estimation of TSR from histopathological images of colorectal cancer. The method is based on convolutional neural networks which were trained to classify colorectal cancer tissue in hematoxylin-eosin stained samples into three classes: stroma, tumor and other. The models were trained using a data set that consists of 1343 whole slide images. Three different training setups were applied with a transfer learning approach using domain-specific data i.e. an external colorectal cancer histopathological data set. The three most accurate models were chosen as a classifier, TSR values were predicted and the results were compared to a visual TSR estimation made by a pathologist. The results suggest that classification accuracy does not improve when domain-specific data are used in the pre-training of the convolutional neural network models in the task at hand. Classification accuracy for stroma, tumor and other reached 96.1$\%$ on an independent test set. Among the three classes the best model gained the highest accuracy (99.3$\%$) for class tumor. When TSR was predicted with the best model, the correlation between the predicted values and values estimated by an experienced pathologist was 0.57. Further research is needed to study associations between computationally predicted TSR values and other clinicopathological factors of colorectal cancer and the overall survival of the patients.
ACROBAT -- a multi-stain breast cancer histological whole-slide-image data set from routine diagnostics for computational pathology
Nov 24, 2022Abstract:The analysis of FFPE tissue sections stained with haematoxylin and eosin (H&E) or immunohistochemistry (IHC) is an essential part of the pathologic assessment of surgically resected breast cancer specimens. IHC staining has been broadly adopted into diagnostic guidelines and routine workflows to manually assess status and scoring of several established biomarkers, including ER, PGR, HER2 and KI67. However, this is a task that can also be facilitated by computational pathology image analysis methods. The research in computational pathology has recently made numerous substantial advances, often based on publicly available whole slide image (WSI) data sets. However, the field is still considerably limited by the sparsity of public data sets. In particular, there are no large, high quality publicly available data sets with WSIs of matching IHC and H&E-stained tissue sections. Here, we publish the currently largest publicly available data set of WSIs of tissue sections from surgical resection specimens from female primary breast cancer patients with matched WSIs of corresponding H&E and IHC-stained tissue, consisting of 4,212 WSIs from 1,153 patients. The primary purpose of the data set was to facilitate the ACROBAT WSI registration challenge, aiming at accurately aligning H&E and IHC images. For research in the area of image registration, automatic quantitative feedback on registration algorithm performance remains available through the ACROBAT challenge website, based on more than 37,000 manually annotated landmark pairs from 13 annotators. Beyond registration, this data set has the potential to enable many different avenues of computational pathology research, including stain-guided learning, virtual staining, unsupervised pre-training, artefact detection and stain-independent models.
Deformation equivariant cross-modality image synthesis with paired non-aligned training data
Aug 26, 2022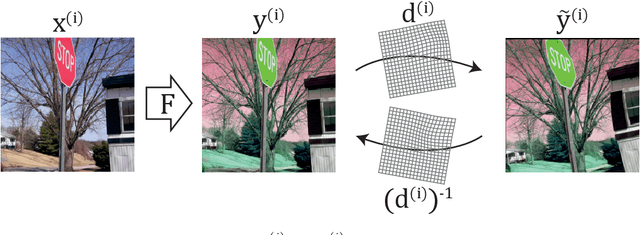
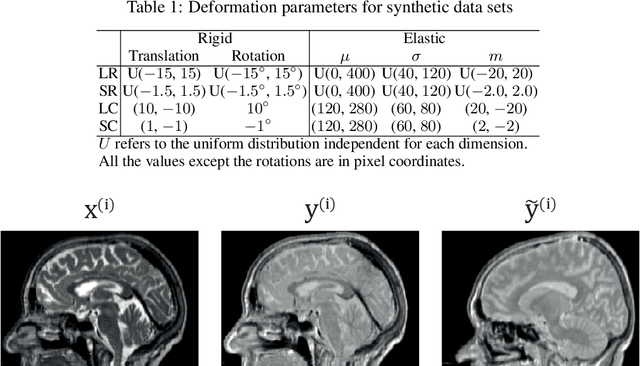
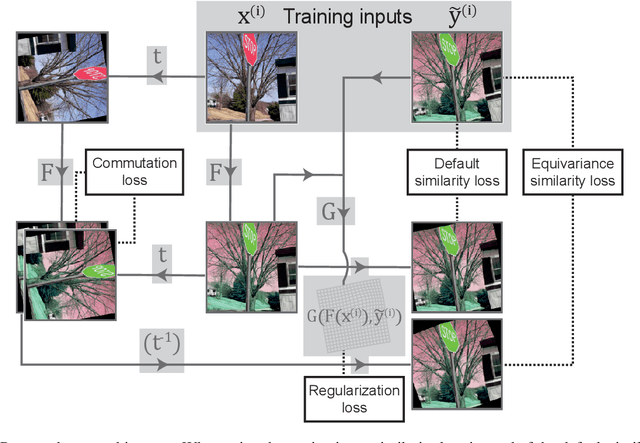
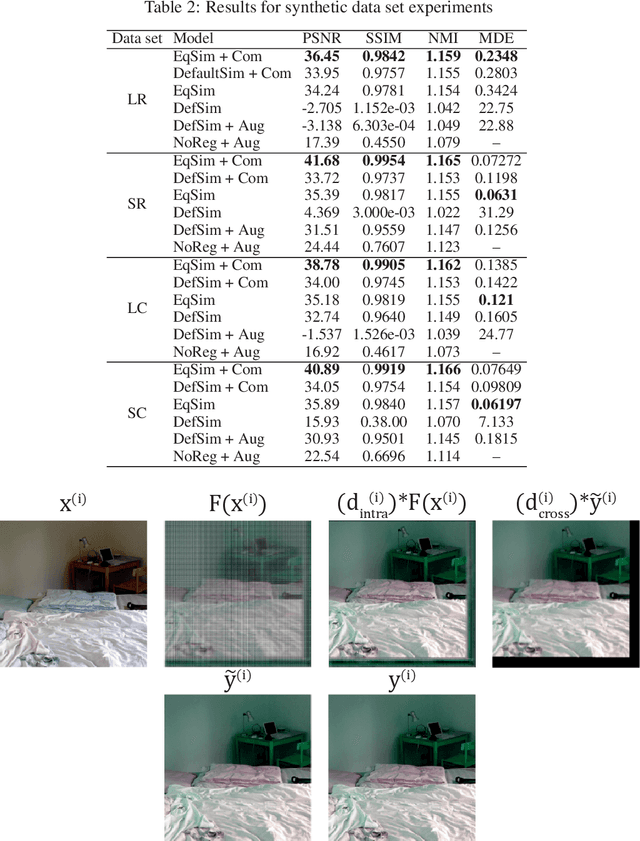
Abstract:Cross-modality image synthesis is an active research topic with multiple medical clinically relevant applications. Recently, methods allowing training with paired but misaligned data have started to emerge. However, no robust and well-performing methods applicable to a wide range of real world data sets exist. In this work, we propose a generic solution to the problem of cross-modality image synthesis with paired but non-aligned data by introducing new deformation equivariance encouraging loss functions. The method consists of joint training of an image synthesis network together with separate registration networks and allows adversarial training conditioned on the input even with misaligned data. The work lowers the bar for new clinical applications by allowing effortless training of cross-modality image synthesis networks for more difficult data sets and opens up opportunities for the development of new generic learning based cross-modality registration algorithms.
Predicting molecular phenotypes from histopathology images: a transcriptome-wide expression-morphology analysis in breast cancer
Sep 18, 2020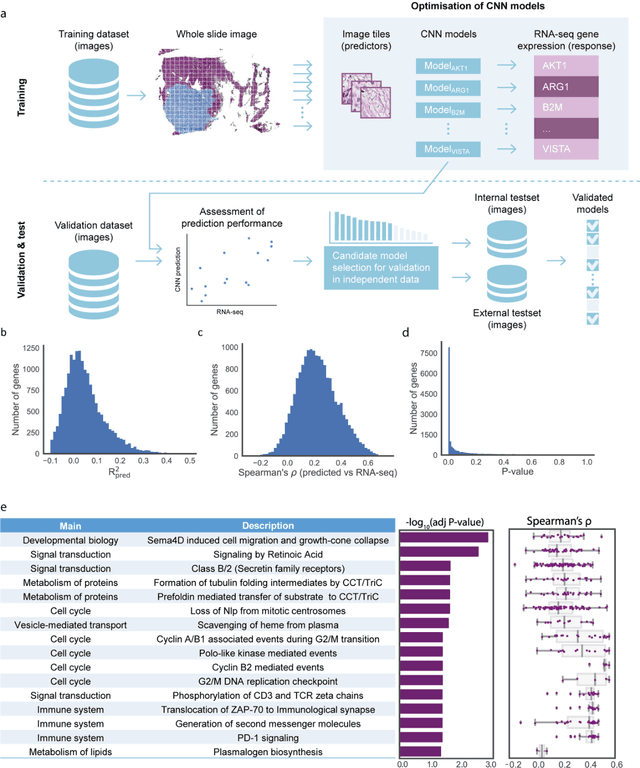
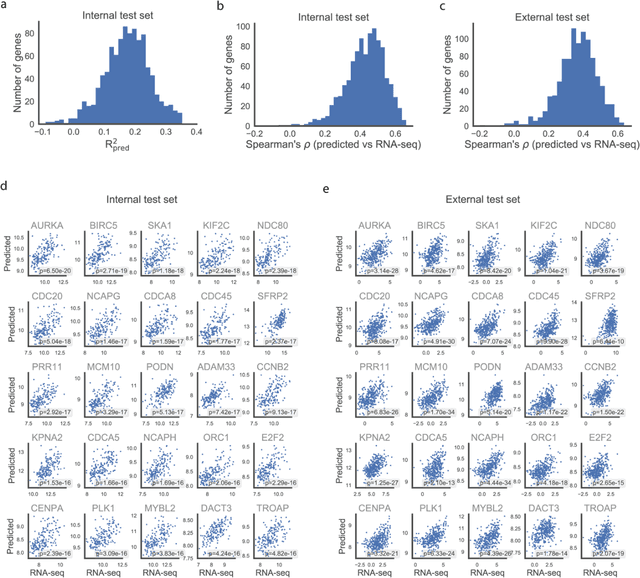
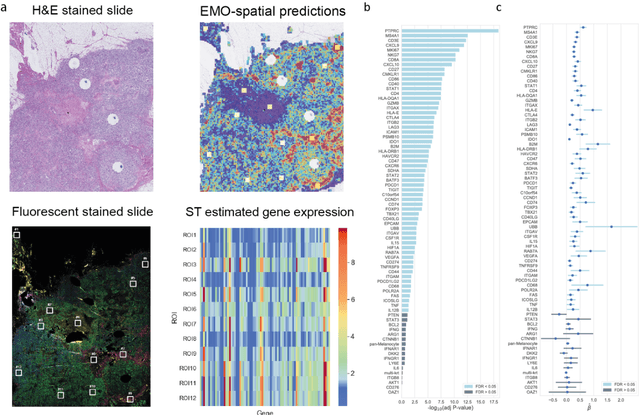
Abstract:Molecular phenotyping is central in cancer precision medicine, but remains costly and standard methods only provide a tumour average profile. Microscopic morphological patterns observable in histopathology sections from tumours are determined by the underlying molecular phenotype and associated with clinical factors. The relationship between morphology and molecular phenotype has a potential to be exploited for prediction of the molecular phenotype from the morphology visible in histopathology images. We report the first transcriptome-wide Expression-MOrphology (EMO) analysis in breast cancer, where gene-specific models were optimised and validated for prediction of mRNA expression both as a tumour average and in spatially resolved manner. Individual deep convolutional neural networks (CNNs) were optimised to predict the expression of 17,695 genes from hematoxylin and eosin (HE) stained whole slide images (WSIs). Predictions for 9,334 (52.75%) genes were significantly associated with RNA-sequencing estimates (FDR adjusted p-value < 0.05). 1,011 of the genes were brought forward for validation, with 876 (87%) and 908 (90%) successfully replicated in internal and external test data, respectively. Predicted spatial intra-tumour variabilities in expression were validated in 76 genes, out of which 59 (77.6%) had a significant association (FDR adjusted p-value < 0.05) with spatial transcriptomics estimates. These results suggest that the proposed methodology can be applied to predict both tumour average gene expression and intra-tumour spatial expression directly from morphology, thus providing a scalable approach to characterise intra-tumour heterogeneity.
Detection of Perineural Invasion in Prostate Needle Biopsies with Deep Neural Networks
Apr 03, 2020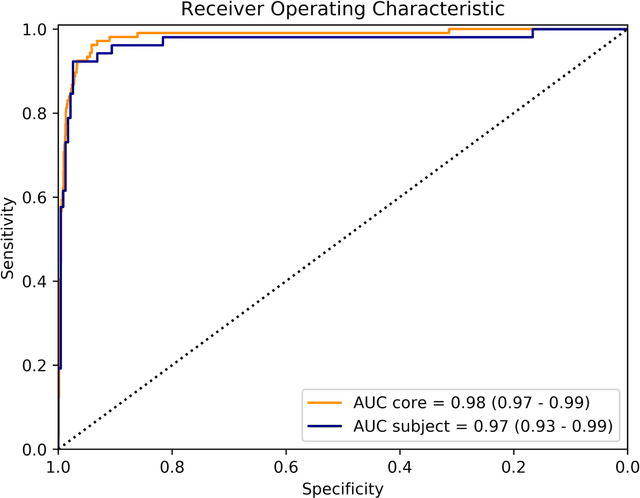
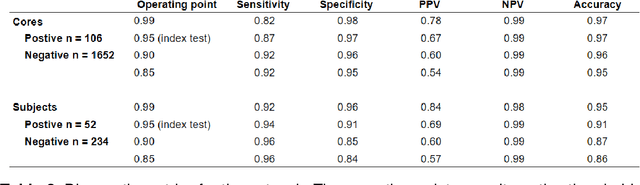
Abstract:Background: The detection of perineural invasion (PNI) by carcinoma in prostate biopsies has been shown to be associated with poor prognosis. The assessment and quantification of PNI is; however, labor intensive. In the study we aimed to develop an algorithm based on deep neural networks to aid pathologists in this task. Methods: We collected, digitized and pixel-wise annotated the PNI findings in each of the approximately 80,000 biopsy cores from the 7,406 men who underwent biopsy in the prospective and diagnostic STHLM3 trial between 2012 and 2014. In total, 485 biopsy cores showed PNI. We also digitized more than 10% (n=8,318) of the PNI negative biopsy cores. Digitized biopsies from a random selection of 80% of the men were used to build deep neural networks, and the remaining 20% were used to evaluate the performance of the algorithm. Results: For the detection of PNI in prostate biopsy cores the network had an estimated area under the receiver operating characteristics curve of 0.98 (95% CI 0.97-0.99) based on 106 PNI positive cores and 1,652 PNI negative cores in the independent test set. For the pre-specified operating point this translates to sensitivity of 0.87 and specificity of 0.97. The corresponding positive and negative predictive values were 0.67 and 0.99, respectively. For localizing the regions of PNI within a slide we estimated an average intersection over union of 0.50 (CI: 0.46-0.55). Conclusion: We have developed an algorithm based on deep neural networks for detecting PNI in prostate biopsies with apparently acceptable diagnostic properties. These algorithms have the potential to aid pathologists in the day-to-day work by drastically reducing the number of biopsy cores that need to be assessed for PNI and by highlighting regions of diagnostic interest.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge