Leena Latonen
Staining normalization in histopathology: Method benchmarking using multicenter dataset
Jun 23, 2025Abstract:Hematoxylin and Eosin (H&E) has been the gold standard in tissue analysis for decades, however, tissue specimens stained in different laboratories vary, often significantly, in appearance. This variation poses a challenge for both pathologists' and AI-based downstream analysis. Minimizing stain variation computationally is an active area of research. To further investigate this problem, we collected a unique multi-center tissue image dataset, wherein tissue samples from colon, kidney, and skin tissue blocks were distributed to 66 different labs for routine H&E staining. To isolate staining variation, other factors affecting the tissue appearance were kept constant. Further, we used this tissue image dataset to compare the performance of eight different stain normalization methods, including four traditional methods, namely, histogram matching, Macenko, Vahadane, and Reinhard normalization, and two deep learning-based methods namely CycleGAN and Pixp2pix, both with two variants each. We used both quantitative and qualitative evaluation to assess the performance of these methods. The dataset's inter-laboratory staining variation could also guide strategies to improve model generalizability through varied training data
The ACROBAT 2022 Challenge: Automatic Registration Of Breast Cancer Tissue
May 29, 2023Abstract:The alignment of tissue between histopathological whole-slide-images (WSI) is crucial for research and clinical applications. Advances in computing, deep learning, and availability of large WSI datasets have revolutionised WSI analysis. Therefore, the current state-of-the-art in WSI registration is unclear. To address this, we conducted the ACROBAT challenge, based on the largest WSI registration dataset to date, including 4,212 WSIs from 1,152 breast cancer patients. The challenge objective was to align WSIs of tissue that was stained with routine diagnostic immunohistochemistry to its H&E-stained counterpart. We compare the performance of eight WSI registration algorithms, including an investigation of the impact of different WSI properties and clinical covariates. We find that conceptually distinct WSI registration methods can lead to highly accurate registration performances and identify covariates that impact performances across methods. These results establish the current state-of-the-art in WSI registration and guide researchers in selecting and developing methods.
ACROBAT -- a multi-stain breast cancer histological whole-slide-image data set from routine diagnostics for computational pathology
Nov 24, 2022Abstract:The analysis of FFPE tissue sections stained with haematoxylin and eosin (H&E) or immunohistochemistry (IHC) is an essential part of the pathologic assessment of surgically resected breast cancer specimens. IHC staining has been broadly adopted into diagnostic guidelines and routine workflows to manually assess status and scoring of several established biomarkers, including ER, PGR, HER2 and KI67. However, this is a task that can also be facilitated by computational pathology image analysis methods. The research in computational pathology has recently made numerous substantial advances, often based on publicly available whole slide image (WSI) data sets. However, the field is still considerably limited by the sparsity of public data sets. In particular, there are no large, high quality publicly available data sets with WSIs of matching IHC and H&E-stained tissue sections. Here, we publish the currently largest publicly available data set of WSIs of tissue sections from surgical resection specimens from female primary breast cancer patients with matched WSIs of corresponding H&E and IHC-stained tissue, consisting of 4,212 WSIs from 1,153 patients. The primary purpose of the data set was to facilitate the ACROBAT WSI registration challenge, aiming at accurately aligning H&E and IHC images. For research in the area of image registration, automatic quantitative feedback on registration algorithm performance remains available through the ACROBAT challenge website, based on more than 37,000 manually annotated landmark pairs from 13 annotators. Beyond registration, this data set has the potential to enable many different avenues of computational pathology research, including stain-guided learning, virtual staining, unsupervised pre-training, artefact detection and stain-independent models.
Deformation equivariant cross-modality image synthesis with paired non-aligned training data
Aug 26, 2022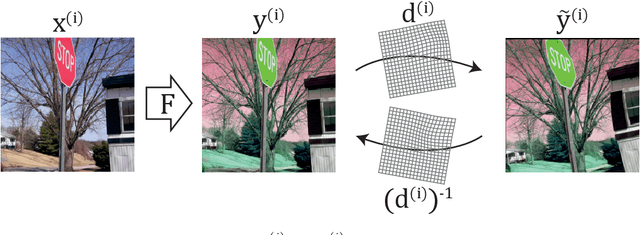
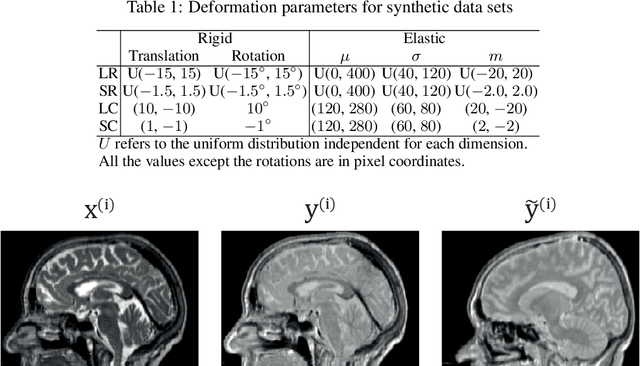
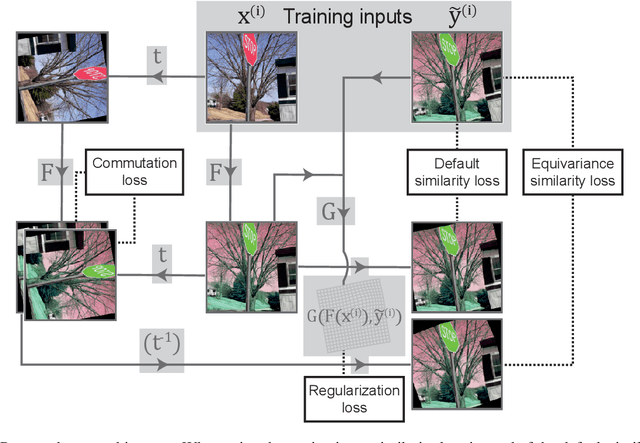
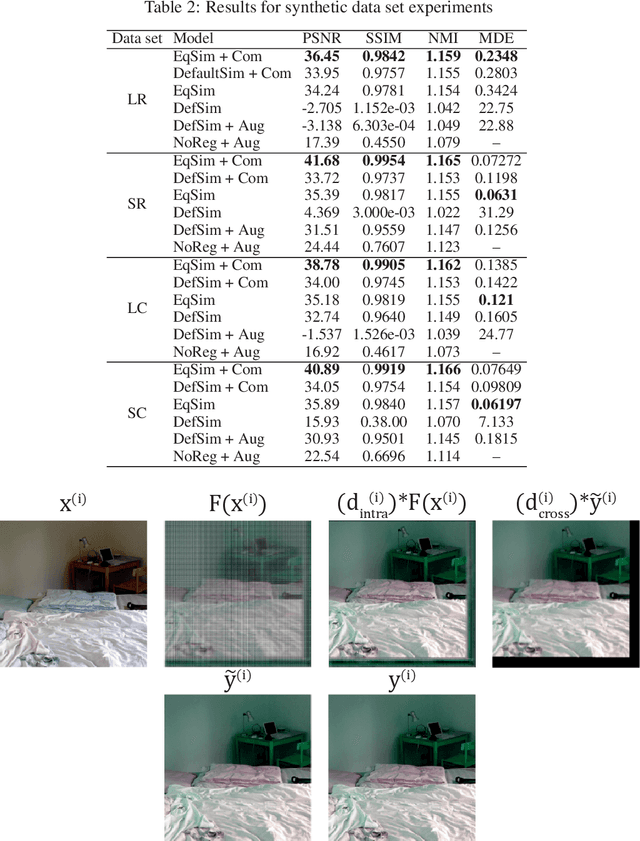
Abstract:Cross-modality image synthesis is an active research topic with multiple medical clinically relevant applications. Recently, methods allowing training with paired but misaligned data have started to emerge. However, no robust and well-performing methods applicable to a wide range of real world data sets exist. In this work, we propose a generic solution to the problem of cross-modality image synthesis with paired but non-aligned data by introducing new deformation equivariance encouraging loss functions. The method consists of joint training of an image synthesis network together with separate registration networks and allows adversarial training conditioned on the input even with misaligned data. The work lowers the bar for new clinical applications by allowing effortless training of cross-modality image synthesis networks for more difficult data sets and opens up opportunities for the development of new generic learning based cross-modality registration algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge