Philippe Weitz
The ACROBAT 2022 Challenge: Automatic Registration Of Breast Cancer Tissue
May 29, 2023Abstract:The alignment of tissue between histopathological whole-slide-images (WSI) is crucial for research and clinical applications. Advances in computing, deep learning, and availability of large WSI datasets have revolutionised WSI analysis. Therefore, the current state-of-the-art in WSI registration is unclear. To address this, we conducted the ACROBAT challenge, based on the largest WSI registration dataset to date, including 4,212 WSIs from 1,152 breast cancer patients. The challenge objective was to align WSIs of tissue that was stained with routine diagnostic immunohistochemistry to its H&E-stained counterpart. We compare the performance of eight WSI registration algorithms, including an investigation of the impact of different WSI properties and clinical covariates. We find that conceptually distinct WSI registration methods can lead to highly accurate registration performances and identify covariates that impact performances across methods. These results establish the current state-of-the-art in WSI registration and guide researchers in selecting and developing methods.
Increasing the usefulness of already existing annotations through WSI registration
Mar 12, 2023Abstract:Computational pathology methods have the potential to improve access to precision medicine, as well as the reproducibility and accuracy of pathological diagnoses. Particularly the analysis of whole-slide-images (WSIs) of immunohistochemically (IHC) stained tissue sections could benefit from computational pathology methods. However, scoring biomarkers such as KI67 in IHC WSIs often necessitates the detection of areas of invasive cancer. Training cancer detection models often requires annotations, which is time-consuming and therefore costly. Currently, cancer regions are typically annotated in WSIs of haematoxylin and eosin (H&E) stained tissue sections. In this study, we investigate the possibility to register annotations that were made in H&E WSIs to their IHC counterparts. Two pathologists annotated regions of invasive cancer in WSIs of 272 breast cancer cases. For each case, a matched H&E and KI67 WSI are available, resulting in 544 WSIs with invasive cancer annotations. We find that cancer detection CNNs that were trained with annotations registered from the H&E to the KI67 WSIs only differ slightly in calibration but not in performance compared to cancer detection models trained on annotations made directly in the KI67 WSIs in a test set consisting of 54 cases. The mean slide-level AUROC is 0.974 [0.964, 0.982] for models trained with the KI67 annotations and 0.974 [0.965, 0.982] for models trained using registered annotations. This indicates that WSI registration has the potential to reduce the need for IHC-specific annotations. This could significantly increase the usefulness of already existing annotations.
ACROBAT -- a multi-stain breast cancer histological whole-slide-image data set from routine diagnostics for computational pathology
Nov 24, 2022Abstract:The analysis of FFPE tissue sections stained with haematoxylin and eosin (H&E) or immunohistochemistry (IHC) is an essential part of the pathologic assessment of surgically resected breast cancer specimens. IHC staining has been broadly adopted into diagnostic guidelines and routine workflows to manually assess status and scoring of several established biomarkers, including ER, PGR, HER2 and KI67. However, this is a task that can also be facilitated by computational pathology image analysis methods. The research in computational pathology has recently made numerous substantial advances, often based on publicly available whole slide image (WSI) data sets. However, the field is still considerably limited by the sparsity of public data sets. In particular, there are no large, high quality publicly available data sets with WSIs of matching IHC and H&E-stained tissue sections. Here, we publish the currently largest publicly available data set of WSIs of tissue sections from surgical resection specimens from female primary breast cancer patients with matched WSIs of corresponding H&E and IHC-stained tissue, consisting of 4,212 WSIs from 1,153 patients. The primary purpose of the data set was to facilitate the ACROBAT WSI registration challenge, aiming at accurately aligning H&E and IHC images. For research in the area of image registration, automatic quantitative feedback on registration algorithm performance remains available through the ACROBAT challenge website, based on more than 37,000 manually annotated landmark pairs from 13 annotators. Beyond registration, this data set has the potential to enable many different avenues of computational pathology research, including stain-guided learning, virtual staining, unsupervised pre-training, artefact detection and stain-independent models.
Using deep learning to detect patients at risk for prostate cancer despite benign biopsies
Jul 31, 2021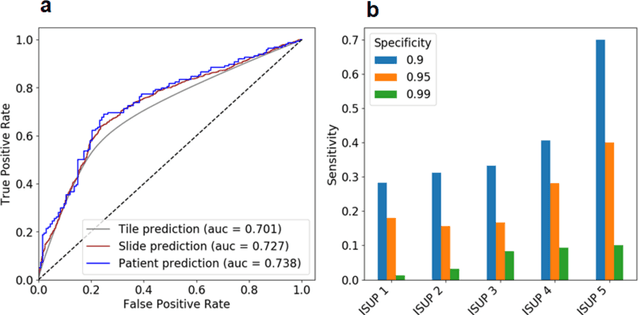
Abstract:Background: Transrectal ultrasound guided systematic biopsies of the prostate is a routine procedure to establish a prostate cancer diagnosis. However, the 10-12 prostate core biopsies only sample a relatively small volume of the prostate, and tumour lesions in regions between biopsy cores can be missed, leading to a well-known low sensitivity to detect clinically relevant cancer. As a proof-of-principle, we developed and validated a deep convolutional neural network model to distinguish between morphological patterns in benign prostate biopsy whole slide images from men with and without established cancer. Methods: This study included 14,354 hematoxylin and eosin stained whole slide images from benign prostate biopsies from 1,508 men in two groups: men without an established prostate cancer (PCa) diagnosis and men with at least one core biopsy diagnosed with PCa. 80% of the participants were assigned as training data and used for model optimization (1,211 men), and the remaining 20% (297 men) as a held-out test set used to evaluate model performance. An ensemble of 10 deep convolutional neural network models was optimized for classification of biopsies from men with and without established cancer. Hyperparameter optimization and model selection was performed by cross-validation in the training data . Results: Area under the receiver operating characteristic curve (ROC-AUC) was estimated as 0.727 (bootstrap 95% CI: 0.708-0.745) on biopsy level and 0.738 (bootstrap 95% CI: 0.682 - 0.796) on man level. At a specificity of 0.9 the model had an estimated sensitivity of 0.348. Conclusion: The developed model has the ability to detect men with risk of missed PCa due to under-sampling of the prostate. The proposed model has the potential to reduce the number of false negative cases in routine systematic prostate biopsies and to indicate men who could benefit from MRI-guided re-biopsy.
Transcriptome-wide prediction of prostate cancer gene expression from histopathology images using co-expression based convolutional neural networks
Apr 19, 2021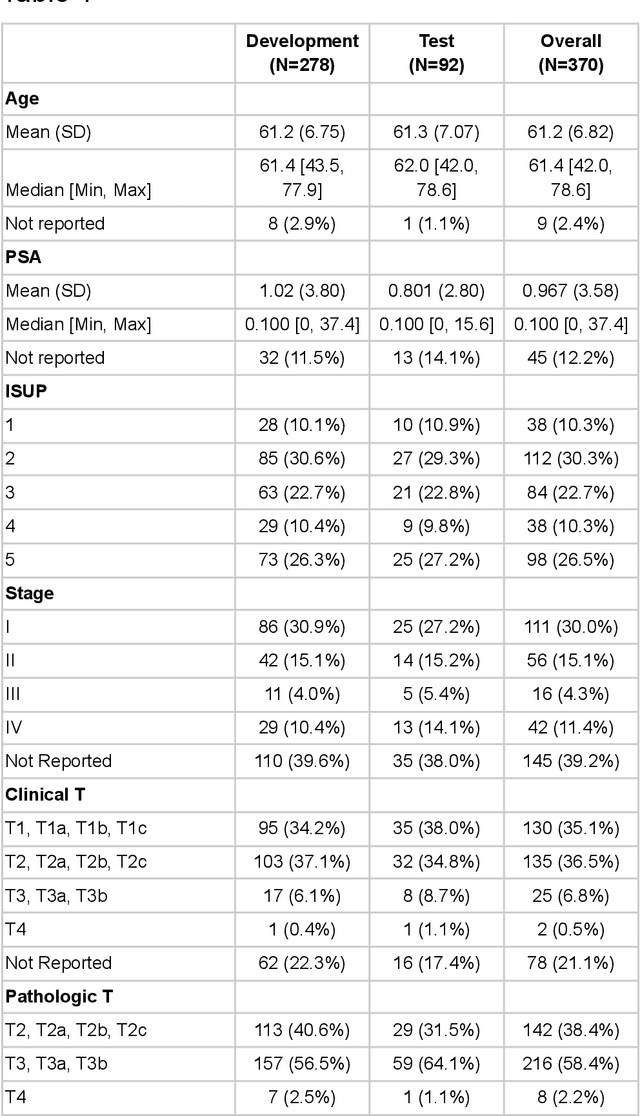
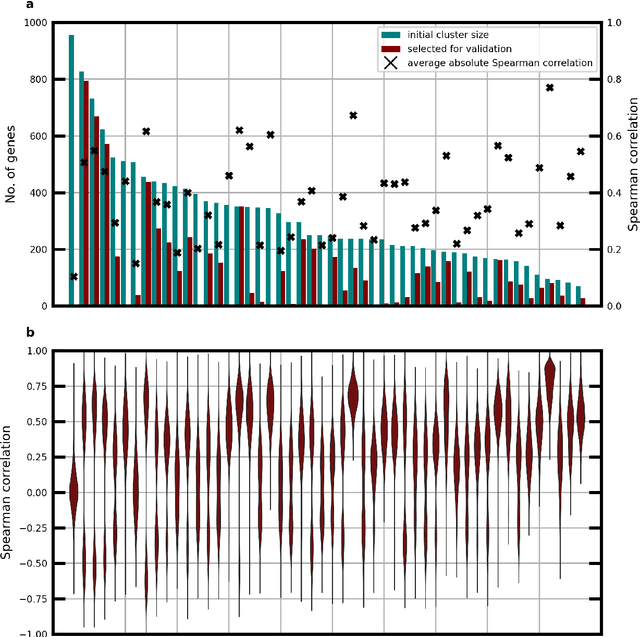
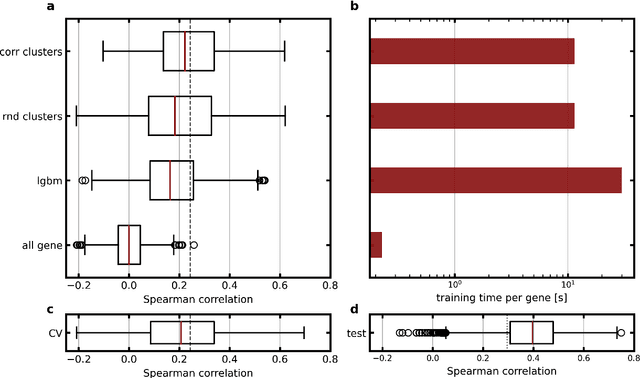
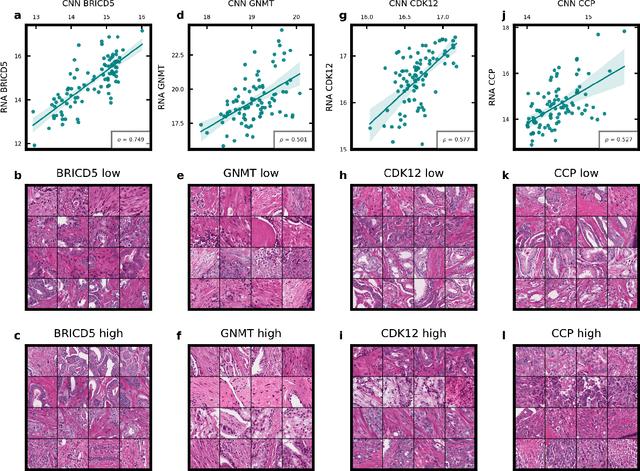
Abstract:Molecular phenotyping by gene expression profiling is common in contemporary cancer research and in molecular diagnostics. However, molecular profiling remains costly and resource intense to implement, and is just starting to be introduced into clinical diagnostics. Molecular changes, including genetic alterations and gene expression changes, occuring in tumors cause morphological changes in tissue, which can be observed on the microscopic level. The relationship between morphological patterns and some of the molecular phenotypes can be exploited to predict molecular phenotypes directly from routine haematoxylin and eosin (H&E) stained whole slide images (WSIs) using deep convolutional neural networks (CNNs). In this study, we propose a new, computationally efficient approach for disease specific modelling of relationships between morphology and gene expression, and we conducted the first transcriptome-wide analysis in prostate cancer, using CNNs to predict bulk RNA-sequencing estimates from WSIs of H&E stained tissue. The work is based on the TCGA PRAD study and includes both WSIs and RNA-seq data for 370 patients. Out of 15586 protein coding and sufficiently frequently expressed transcripts, 6618 had predicted expression significantly associated with RNA-seq estimates (FDR-adjusted p-value < 1*10-4) in a cross-validation. 5419 (81.9%) of these were subsequently validated in a held-out test set. We also demonstrate the ability to predict a prostate cancer specific cell cycle progression score directly from WSIs. These findings suggest that contemporary computer vision models offer an inexpensive and scalable solution for prediction of gene expression phenotypes directly from WSIs, providing opportunity for cost-effective large-scale research studies and molecular diagnostics.
Radiomic Feature Stability Analysis based on Probabilistic Segmentations
Oct 13, 2019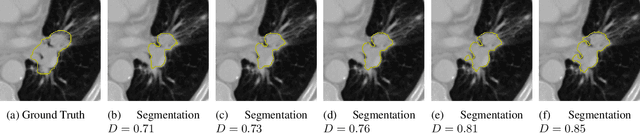
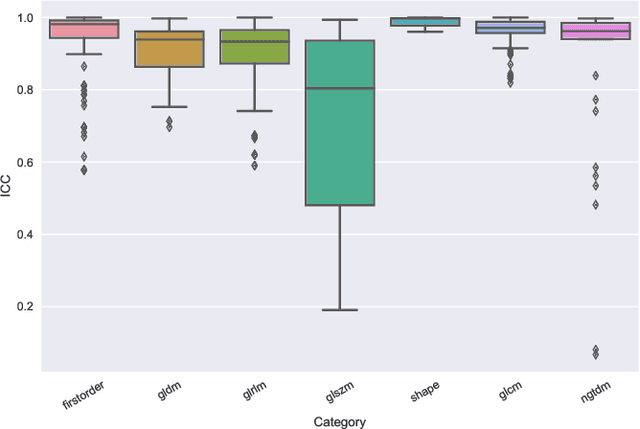
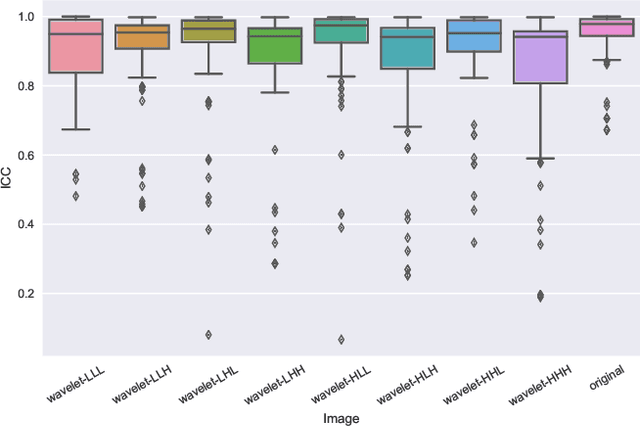
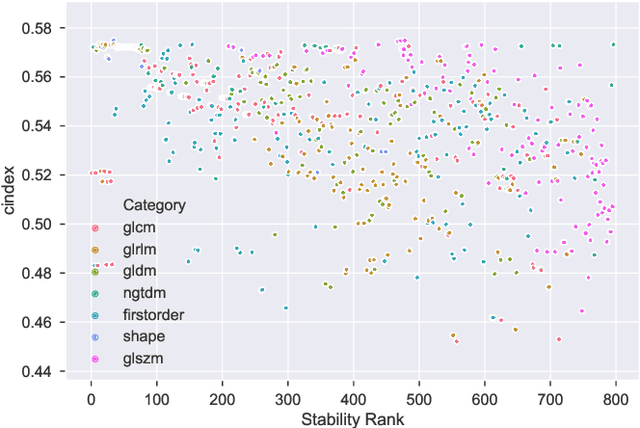
Abstract:Identifying image features that are robust with respect to segmentation variability and domain shift is a tough challenge in radiomics. So far, this problem has mainly been tackled in test-retest analyses. In this work we analyze radiomics feature stability based on probabilistic automated segmentation hypotheses. Based on a public lung cancer dataset, we generate an arbitrary number of plausible segmentations using a Probabilistic U-Net. From these segmentations, we extract a high number of plausible feature vectors for each lung tumor and analyze feature variance with respect to the segmentations. Our results suggest that there are groups of radiomic features that are more (e.g. statistics features) and less (e.g. gray-level size zone matrix features) robust against segmentation variability. Finally, we demonstrate that segmentation variance impacts the performance of a prognostic lung cancer survival model and propose a new and potentially more robust radiomics feature selection workflow.
Image-based Survival Analysis for Lung Cancer Patients using CNNs
Oct 08, 2018
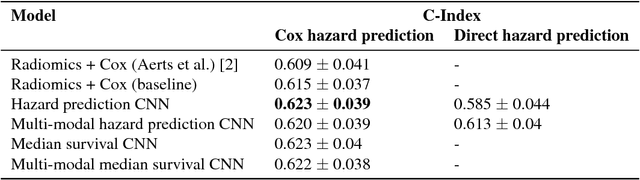
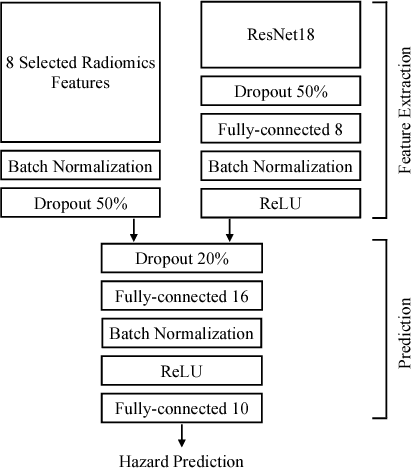
Abstract:Traditional survival models such as the Cox proportional hazards model are typically based on scalar or categorical clinical features. With the advent of increasingly large image datasets, it has become feasible to incorporate quantitative image features into survival prediction. So far, this kind of analysis is mostly based on radiomics features, i.e. a fixed set of features that is mathematically defined a priori. To capture highly abstract information, it is desirable to learn the feature extraction using convolutional neural networks. However, for tomographic medical images, model training is difficult because on the one hand, only few samples of 3D image data fit into one batch at once and on the other hand, survival loss functions are essentially ordering measures that require large batch sizes. In this work, we show that by simplifying survival analysis to median survival classification, convolutional neural networks can be trained with small batch sizes and learn features that predict survival equally well as end-to-end hazard prediction networks. Our approach outperforms the previous state of the art in a publicly available lung cancer dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge