Pascal Spincemaille
QSM-RimDS: A highly sensitive paramagnetic rim lesion detection and segmentation tool for multiple sclerosis lesions
Dec 13, 2024



Abstract:Paramagnetic rim lesions (PRLs) are imaging biomarker of the innate immune response in MS lesions. QSM-RimNet, a state-of-the-art tool for PRLs detection on QSM, can identify PRLs but requires precise QSM lesion mask and does not provide rim segmentation. Therefore, the aims of this study are to develop QSM-RimDS algorithm to detect PRLs using the readily available FLAIR lesion mask and to provide rim segmentation for microglial quantification. QSM-RimDS, a deep-learning based tool for joint PRL rim segmentation and PRL detection has been developed. QSM-RimDS has obtained state-of-the art performance in PRL detection and therefore has the potential to be used in clinical practice as a tool to assist human readers for the time-consuming PRL detection and segmentation task. QSM-RimDS is made publicly available [https://github.com/kennyha85/QSM_RimDS]
Implicit neural representation for free-breathing MR fingerprinting (INR-MRF): co-registered 3D whole-liver water T1, water T2, proton density fat fraction, and R2* mapping
Oct 19, 2024



Abstract:Purpose: To develop an MRI technique for free-breathing 3D whole-liver quantification of water T1, water T2, proton density fat fraction (PDFF), R2*. Methods: An Eight-echo spoiled gradient echo pulse sequence with spiral readout was developed by interleaving inversion recovery and T2 magnetization preparation. We propose a neural network based on a 4D and a 3D implicit neural representation (INR) which simultaneously learns the motion deformation fields and the static reference frame MRI subspace images respectively. Water and fat singular images were separated during network training, with no need of performing retrospective water-fat separation. T1, T2, R2* and proton density fat fraction (PDFF) produced by the proposed method were validated in vivo on 10 healthy subjects, using quantitative maps generated from conventional scans as reference. Results: Our results showed minimal bias and narrow 95% limits of agreement on T1, T2, R2* and PDFF values in the liver compared to conventional breath-holding scans. Conclusions: INR-MRF enabled co-registered 3D whole liver T1, T2, R2* and PDFF mapping in a single free-breathing scan.
MRI quantification of liver fibrosis using diamagnetic susceptibility: An ex-vivo feasibility study
Oct 04, 2024



Abstract:In chronic liver disease, liver fibrosis develops as excessive deposition of extracellular matrix macromolecules, predominantly collagens, progressively form fibrous scars that disrupt the hepatic architecture, and fibrosis, iron, and fat are interrelated. Fibrosis is the best predictor of morbidity and mortality in chronic liver disease but liver biopsy, the reference method for diagnosis and staging, is invasive and limited by sampling and interobserver variability and risks of complications. The overall objective of this study was to develop a new non-invasive method to quantify fibrosis using diamagnetic susceptibility sources with histology validation in ex vivo liver explants.
Deep learning improved autofocus for motion artifact reduction and its application in quantitative susceptibility mapping
May 26, 2024Abstract:Purpose: To develop a pipeline for motion artifact correction in mGRE and quantitative susceptibility mapping (QSM). Methods: Deep learning is integrated with autofocus to improve motion artifact suppression, which is applied QSM of patients with Parkinson's disease (PD). The estimation of affine motion parameters in the autofocus method depends on signal-to-noise ratio and lacks accuracy when data sampling occurs outside the k-space center. A deep learning strategy is employed to remove the residual motion artifacts in autofocus. Results: Results obtained in simulated brain data (n =15) with reference truth show that the proposed autofocus deep learning method significantly improves the image quality of mGRE and QSM (p = 0.001 for SSIM, p < 0.0001 for PSNR and RMSE). Results from 10 PD patients with real motion artifacts in QSM have also been corrected using the proposed method and sent to an experienced radiologist for image quality evaluation, and the average image quality score has increased (p=0.0039). Conclusions: The proposed method enables substantial suppression of motion artifacts in mGRE and QSM.
Multi-delay arterial spin-labeled perfusion estimation with biophysics simulation and deep learning
Nov 17, 2023



Abstract:Purpose: To develop biophysics-based method for estimating perfusion Q from arterial spin labeling (ASL) images using deep learning. Methods: A 3D U-Net (QTMnet) was trained to estimate perfusion from 4D tracer propagation images. The network was trained and tested on simulated 4D tracer concentration data based on artificial vasculature structure generated by constrained constructive optimization (CCO) method. The trained network was further tested in a synthetic brain ASL image based on vasculature network extracted from magnetic resonance (MR) angiography. The estimations from both trained network and a conventional kinetic model were compared in ASL images acquired from eight healthy volunteers. Results: QTMnet accurately reconstructed perfusion Q from concentration data. Relative error of the synthetic brain ASL image was 7.04% for perfusion Q, lower than the error using single-delay ASL model: 25.15% for Q, and multi-delay ASL model: 12.62% for perfusion Q. Conclusion: QTMnet provides accurate estimation on perfusion parameters and is a promising approach as a clinical ASL MRI image processing pipeline.
Physics-based network fine-tuning for robust quantitative susceptibility mapping from high-pass filtered phase
May 05, 2023



Abstract:Purpose: To improve the generalization ability of convolutional neural network (CNN) based prediction of quantitative susceptibility mapping (QSM) from high-pass filtered phase (HPFP) image. Methods: The proposed network addresses two common generalization issues that arise when using a pre-trained network to predict QSM from HPFP: a) data with unseen voxel sizes, and b) data with unknown high-pass filter parameters. A network fine-tuning step based on a high-pass filtering dipole convolution forward model is proposed to reduce the generalization error of the pre-trained network. A progressive Unet architecture is proposed to improve prediction accuracy without increasing fine-tuning computational cost. Results: In retrospective studies using RMSE, PSNR, SSIM and HFEN as quality metrics, the performance of both Unet and progressive Unet was improved after physics-based fine-tuning at all voxel sizes and most high-pass filtering cutoff frequencies tested in the experiment. Progressive Unet slightly outperformed Unet both before and after fine-tuning. In a prospective study, image sharpness was improved after physics-based fine-tuning for both Unet and progressive Unet. Compared to Unet, progressive Unet had better agreement of regional susceptibility values with reference QSM. Conclusion: The proposed method shows improved robustness compared to the pre-trained network without fine-tuning when the test dataset deviates from training. Our code is available at https://github.com/Jinwei1209/SWI_to_QSM/
Maximum Spherical Mean Value (mSMV) Filtering for Whole Brain Quantitative Susceptibility Mapping
Apr 22, 2023



Abstract:To develop a tissue field filtering algorithm, called maximum Spherical Mean Value (mSMV), for reducing shadow artifacts in quantitative susceptibility mapping (QSM) of the brain without requiring brain tissue erosion. Residual background field is a major source of shadow artifacts in QSM. The mSMV algorithm filters large field values near the border, where the maximum value of the harmonic background field is located. The effectiveness of mSMV for artifact removal was evaluated by comparing with existing QSM algorithms in a simulated numerical brain, 11 healthy volunteers, by assessing image quality in routine clinical patient study $(n=43)$, and by measuring lesion susceptibility values in multiple sclerosis patients $(n=50)$, a total of $n=93$ patients. Numerical simulation showed that mSMV reduces shadow artifacts and improves QSM accuracy. Better shadow reduction, as demonstrated by lower QSM variation in the gray matter and higher QSM image quality score, was also observed in healthy subjects and in patients with hemorrhages, stroke and multiple sclerosis. The mSMV algorithm allows reconstruction of QSMs that are equivalent to those obtained using SMV-filtered dipole inversion without eroding the volume of interest.
mcLARO: Multi-Contrast Learned Acquisition and Reconstruction Optimization for simultaneous quantitative multi-parametric mapping
Apr 07, 2023Abstract:Purpose: To develop a method for rapid sub-millimeter T1, T2, T2* and QSM mapping in a single scan using multi-contrast Learned Acquisition and Reconstruction Optimization (mcLARO). Methods: A pulse sequence was developed by interleaving inversion recovery and T2 magnetization preparations and single-echo and multi-echo gradient echo acquisitions, which sensitized k-space data to T1, T2, T2* and magnetic susceptibility. The proposed mcLARO used a deep learning framework to optimize both the multi-contrast k-space under-sampling pattern and the image reconstruction based on image feature fusion. The proposed mcLARO method with R=8 under-sampling was validated in a retrospective ablation study using fully sampled data as reference and evaluated in a prospective study using separately acquired conventionally sampled quantitative maps as reference standard. Results: The retrospective ablation study showed improved image sharpness of mcLARO compared to the baseline network without multi-contrast sampling pattern optimization or image feature fusion, and negligible bias and narrow 95% limits of agreement on regional T1, T2, T2* and QSM values were obtained by the under-sampled reconstructions compared to the fully sampled reconstruction. The prospective study showed small or negligible bias and narrow 95% limits of agreement on regional T1, T2, T2* and QSM values by mcLARO (5:39 mins) compared to reference scans (40:03 mins in total). Conclusion: mcLARO enabled fast sub-millimeter T1, T2, T2* and QSM mapping in a single scan.
LARO: Learned Acquisition and Reconstruction Optimization to accelerate Quantitative Susceptibility Mapping
Nov 01, 2022Abstract:Quantitative susceptibility mapping (QSM) involves acquisition and reconstruction of a series of images at multi-echo time points to estimate tissue field, which prolongs scan time and requires specific reconstruction technique. In this paper, we present our new framework, called Learned Acquisition and Reconstruction Optimization (LARO), which aims to accelerate the multi-echo gradient echo (mGRE) pulse sequence for QSM. Our approach involves optimizing a Cartesian multi-echo k-space sampling pattern with a deep reconstruction network. Next, this optimized sampling pattern was implemented in an mGRE sequence using Cartesian fan-beam k-space segmenting and ordering for prospective scans. Furthermore, we propose to insert a recurrent temporal feature fusion module into the reconstruction network to capture signal redundancies along echo time. Our ablation studies show that both the optimized sampling pattern and proposed reconstruction strategy help improve the quality of the multi-echo image reconstructions. Generalization experiments show that LARO is robust on the test data with new pathologies and different sequence parameters. Our code is available at https://github.com/Jinwei1209/LARO.git.
Motion Artifact Reduction in Quantitative Susceptibility Mapping using Deep Neural Network
May 04, 2021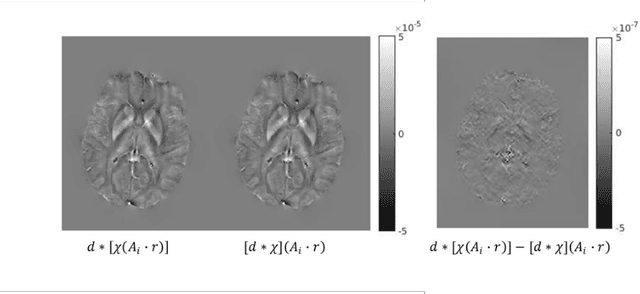
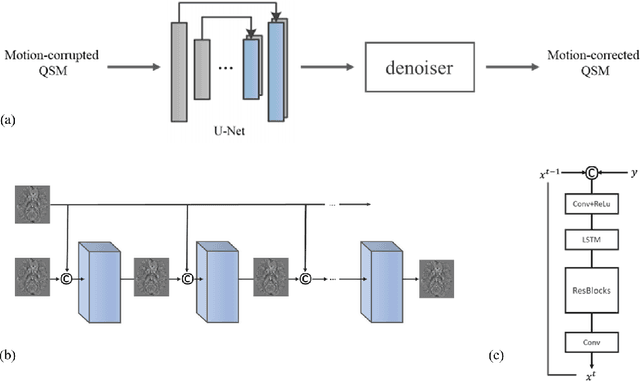
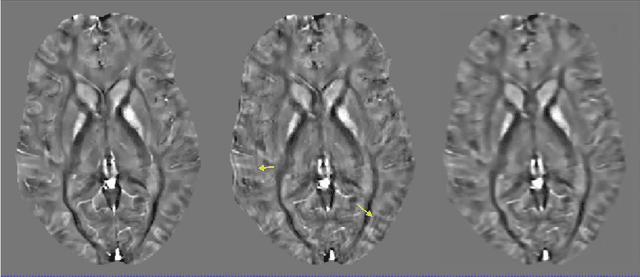
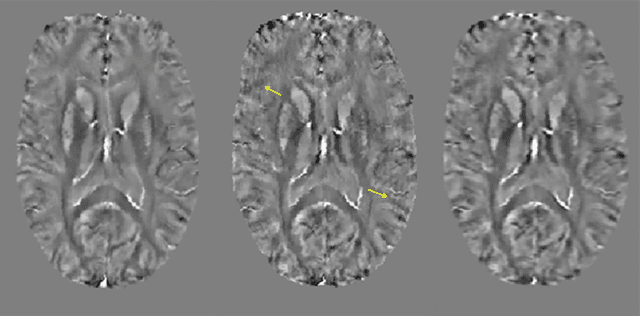
Abstract:An approach to reduce motion artifacts in Quantitative Susceptibility Mapping using deep learning is proposed. We use an affine motion model with randomly created motion profiles to simulate motion-corrupted QSM images. The simulated QSM image is paired with its motion-free reference to train a neural network using supervised learning. The trained network is tested on unseen simulated motion-corrupted QSM images, in healthy volunteers and in Parkinson's disease patients. The results show that motion artifacts, such as ringing and ghosting, were successfully suppressed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge