Nico Karssemeijer
Improving Lesion Volume Measurements on Digital Mammograms
Aug 28, 2023Abstract:Lesion volume is an important predictor for prognosis in breast cancer. We make a step towards a more accurate lesion volume measurement on digital mammograms by developing a model that allows to estimate lesion volumes on processed mammograms, which are the images routinely used by radiologists in clinical practice as well as in breast cancer screening and are available in medical centers. Processed mammograms are obtained from raw mammograms, which are the X-ray data coming directly from the scanner, by applying certain vendor-specific non-linear transformations. At the core of our volume estimation method is a physics-based algorithm for measuring lesion volumes on raw mammograms. We subsequently extend this algorithm to processed mammograms via a deep learning image-to-image translation model that produces synthetic raw mammograms from processed mammograms in a multi-vendor setting. We assess the reliability and validity of our method using a dataset of 1778 mammograms with an annotated mass. Firstly, we investigate the correlations between lesion volumes computed from mediolateral oblique and craniocaudal views, with a resulting Pearson correlation of 0.93 [95% confidence interval (CI) 0.92 - 0.93]. Secondly, we compare the resulting lesion volumes from true and synthetic raw data, with a resulting Pearson correlation of 0.998 [95% CI 0.998 - 0.998] . Finally, for a subset of 100 mammograms with a malign mass and concurrent MRI examination available, we analyze the agreement between lesion volume on mammography and MRI, resulting in an intraclass correlation coefficient of 0.81 [95% CI 0.73 - 0.87] for consistency and 0.78 [95% CI 0.66 - 0.86] for absolute agreement. In conclusion, we developed an algorithm to measure mammographic lesion volume that reached excellent reliability and good validity, when using MRI as ground truth.
Robust Cross-vendor Mammographic Texture Models Using Augmentation-based Domain Adaptation for Long-term Breast Cancer Risk
Jan 10, 2023Abstract:Purpose: Risk-stratified breast cancer screening might improve early detection and efficiency without comprising quality. However, modern mammography-based risk models do not ensure adaptation across vendor-domains and rely on cancer precursors, associated with short-term risk, which might limit long-term risk assessment. We report a cross-vendor mammographic texture model for long-term risk. Approach: The texture model was robustly trained using two systematically designed case-control datasets. Textural features, indicative of future breast cancer, were learned by excluding samples with diagnosed/potential malignancies from training. An augmentation-based domain adaption technique, based on flavorization of mammographic views, ensured generalization across vendor-domains. The model was validated in 66,607 consecutively screened Danish women with flavorized Siemens views and 25,706 Dutch women with Hologic-processed views. Performances were evaluated for interval cancers (IC) within two years from screening and long-term cancers (LTC) from two years after screening. The texture model was combined with established risk factors to flag 10% of women with the highest risk. Results: In Danish women, the texture model achieved an area under the receiver operating characteristic (AUC) of 0.71 and 0.65 for ICs and LTCs, respectively. In Dutch women with Hologic-processed views, the AUCs were not different from AUCs in Danish women with flavorized views. The AUC for texture combined with established risk factors increased to 0.68 for LTCs. The 10% of women flagged as high-risk accounted for 25.5% of ICs and 24.8% of LTCs. Conclusions: The texture model robustly estimated long-term breast cancer risk while adapting to an unseen processed vendor-domain and identified a clinically relevant high-risk subgroup.
Whole-Slide Mitosis Detection in H&E Breast Histology Using PHH3 as a Reference to Train Distilled Stain-Invariant Convolutional Networks
Aug 17, 2018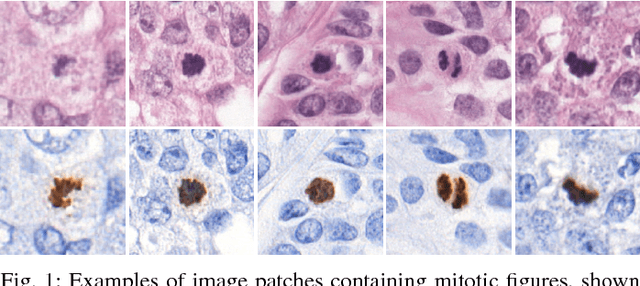
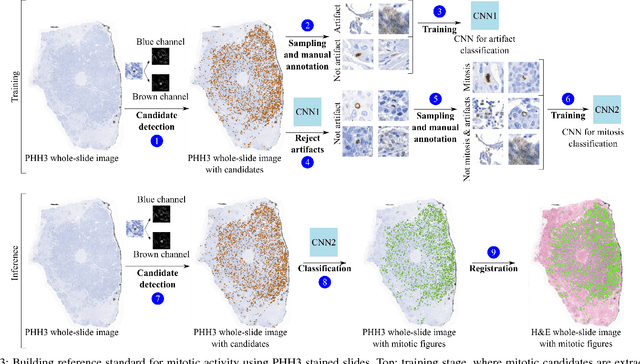
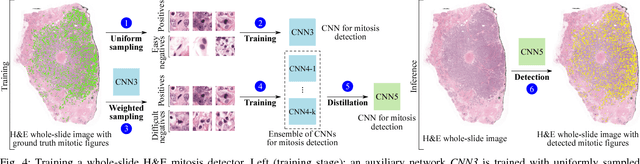
Abstract:Manual counting of mitotic tumor cells in tissue sections constitutes one of the strongest prognostic markers for breast cancer. This procedure, however, is time-consuming and error-prone. We developed a method to automatically detect mitotic figures in breast cancer tissue sections based on convolutional neural networks (CNNs). Application of CNNs to hematoxylin and eosin (H&E) stained histological tissue sections is hampered by: (1) noisy and expensive reference standards established by pathologists, (2) lack of generalization due to staining variation across laboratories, and (3) high computational requirements needed to process gigapixel whole-slide images (WSIs). In this paper, we present a method to train and evaluate CNNs to specifically solve these issues in the context of mitosis detection in breast cancer WSIs. First, by combining image analysis of mitotic activity in phosphohistone-H3 (PHH3) restained slides and registration, we built a reference standard for mitosis detection in entire H&E WSIs requiring minimal manual annotation effort. Second, we designed a data augmentation strategy that creates diverse and realistic H&E stain variations by modifying the hematoxylin and eosin color channels directly. Using it during training combined with network ensembling resulted in a stain invariant mitosis detector. Third, we applied knowledge distillation to reduce the computational requirements of the mitosis detection ensemble with a negligible loss of performance. The system was trained in a single-center cohort and evaluated in an independent multicenter cohort from The Cancer Genome Atlas on the three tasks of the Tumor Proliferation Assessment Challenge (TUPAC). We obtained a performance within the top-3 best methods for most of the tasks of the challenge.
Student Beats the Teacher: Deep Neural Networks for Lateral Ventricles Segmentation in Brain MR
Mar 03, 2018Abstract:Ventricular volume and its progression are known to be linked to several brain diseases such as dementia and schizophrenia. Therefore accurate measurement of ventricle volume is vital for longitudinal studies on these disorders, making automated ventricle segmentation algorithms desirable. In the past few years, deep neural networks have shown to outperform the classical models in many imaging domains. However, the success of deep networks is dependent on manually labeled data sets, which are expensive to acquire especially for higher dimensional data in the medical domain. In this work, we show that deep neural networks can be trained on much-cheaper-to-acquire pseudo-labels (e.g., generated by other automated less accurate methods) and still produce more accurate segmentations compared to the quality of the labels. To show this, we use noisy segmentation labels generated by a conventional region growing algorithm to train a deep network for lateral ventricle segmentation. Then on a large manually annotated test set, we show that the network significantly outperforms the conventional region growing algorithm which was used to produce the training labels for the network. Our experiments report a Dice Similarity Coefficient (DSC) of $0.874$ for the trained network compared to $0.754$ for the conventional region growing algorithm ($p < 0.001$).
* 7 pages, 4 figures, SPIE Medical Imaging 2018 Conference paper
Classifying Symmetrical Differences and Temporal Change in Mammography Using Deep Neural Networks
Aug 01, 2017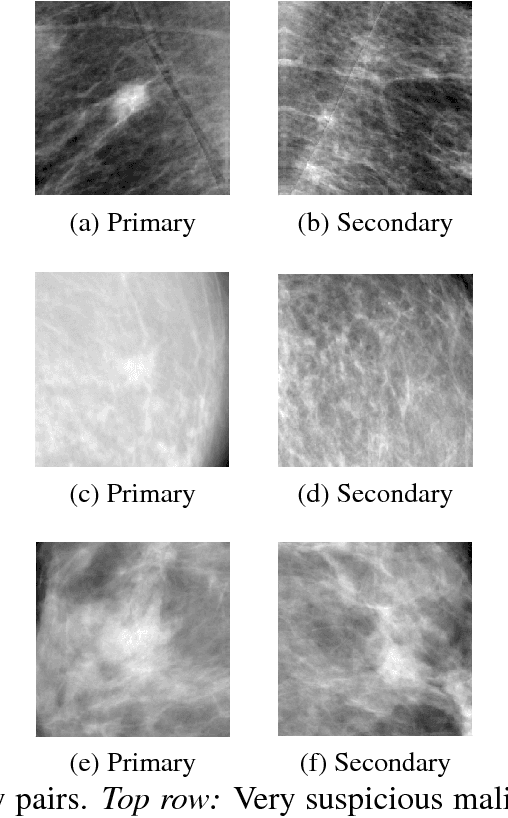

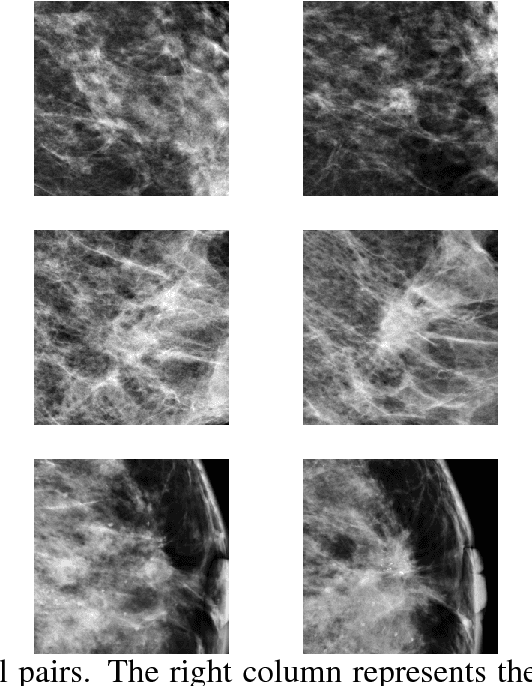
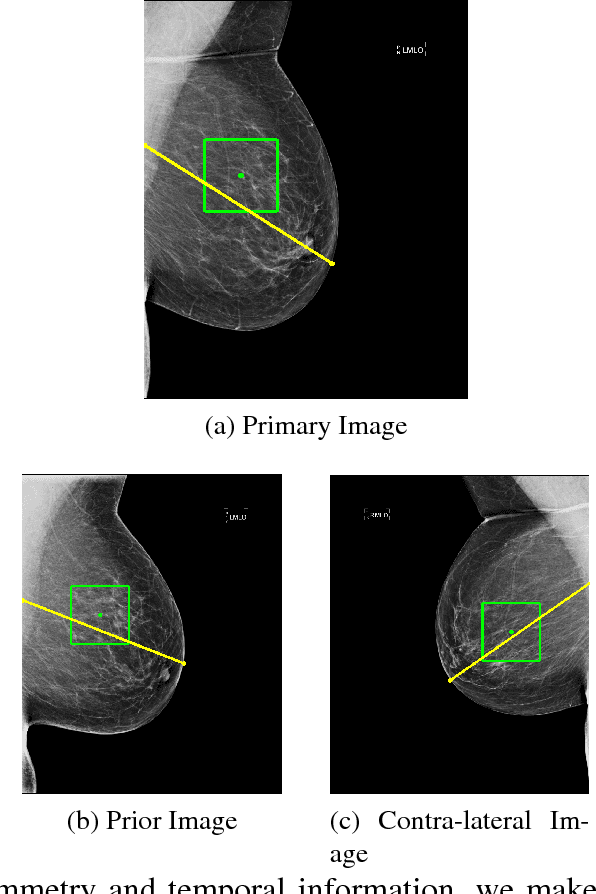
Abstract:We investigate the addition of symmetry and temporal context information to a deep Convolutional Neural Network (CNN) with the purpose of detecting malignant soft tissue lesions in mammography. We employ a simple linear mapping that takes the location of a mass candidate and maps it to either the contra-lateral or prior mammogram and Regions Of Interest (ROI) are extracted around each location. We subsequently explore two different architectures (1) a fusion model employing two datastreams were both ROIs are fed to the network during training and testing and (2) a stage-wise approach where a single ROI CNN is trained on the primary image and subsequently used as feature extractor for both primary and symmetrical or prior ROIs. A 'shallow' Gradient Boosted Tree (GBT) classifier is then trained on the concatenation of these features and used to classify the joint representation. Results shown a significant increase in performance using the first architecture and symmetry information, but only marginal gains in performance using temporal data and the other setting. We feel results are promising and can greatly be improved when more temporal data becomes available.
Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images
May 10, 2017Abstract:Automated classification of histopathological whole-slide images (WSI) of breast tissue requires analysis at very high resolutions with a large contextual area. In this paper, we present context-aware stacked convolutional neural networks (CNN) for classification of breast WSIs into normal/benign, ductal carcinoma in situ (DCIS), and invasive ductal carcinoma (IDC). We first train a CNN using high pixel resolution patches to capture cellular level information. The feature responses generated by this model are then fed as input to a second CNN, stacked on top of the first. Training of this stacked architecture with large input patches enables learning of fine-grained (cellular) details and global interdependence of tissue structures. Our system is trained and evaluated on a dataset containing 221 WSIs of H&E stained breast tissue specimens. The system achieves an AUC of 0.962 for the binary classification of non-malignant and malignant slides and obtains a three class accuracy of 81.3% for classification of WSIs into normal/benign, DCIS, and IDC, demonstrating its potentials for routine diagnostics.
Transfer Learning for Domain Adaptation in MRI: Application in Brain Lesion Segmentation
Feb 25, 2017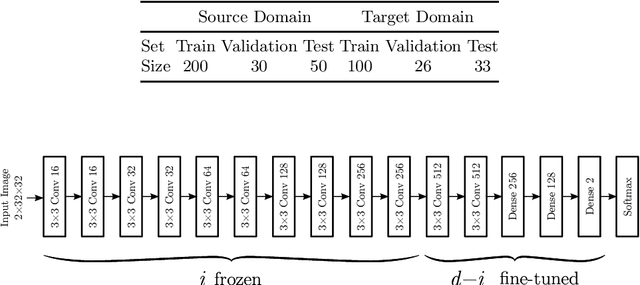
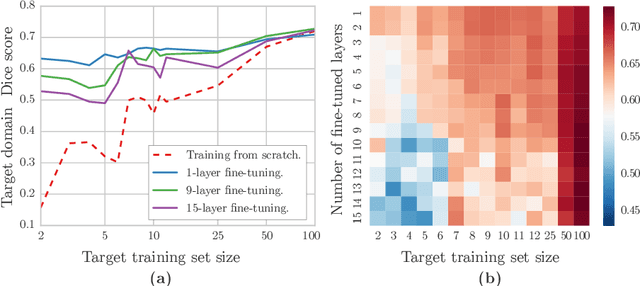
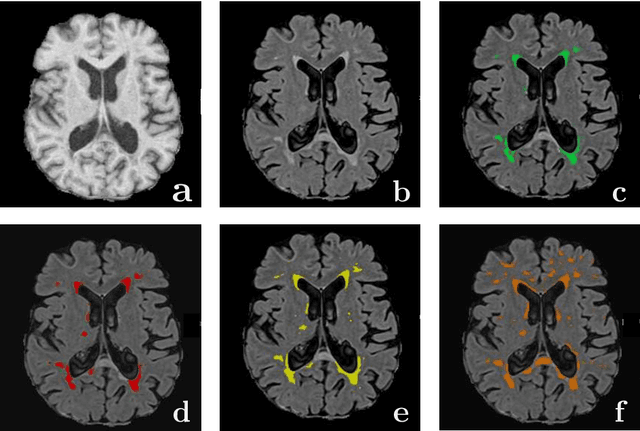
Abstract:Magnetic Resonance Imaging (MRI) is widely used in routine clinical diagnosis and treatment. However, variations in MRI acquisition protocols result in different appearances of normal and diseased tissue in the images. Convolutional neural networks (CNNs), which have shown to be successful in many medical image analysis tasks, are typically sensitive to the variations in imaging protocols. Therefore, in many cases, networks trained on data acquired with one MRI protocol, do not perform satisfactorily on data acquired with different protocols. This limits the use of models trained with large annotated legacy datasets on a new dataset with a different domain which is often a recurring situation in clinical settings. In this study, we aim to answer the following central questions regarding domain adaptation in medical image analysis: Given a fitted legacy model, 1) How much data from the new domain is required for a decent adaptation of the original network?; and, 2) What portion of the pre-trained model parameters should be retrained given a certain number of the new domain training samples? To address these questions, we conducted extensive experiments in white matter hyperintensity segmentation task. We trained a CNN on legacy MR images of brain and evaluated the performance of the domain-adapted network on the same task with images from a different domain. We then compared the performance of the model to the surrogate scenarios where either the same trained network is used or a new network is trained from scratch on the new dataset.The domain-adapted network tuned only by two training examples achieved a Dice score of 0.63 substantially outperforming a similar network trained on the same set of examples from scratch.
* 8 pages, 3 figures
Deep learning-based assessment of tumor-associated stroma for diagnosing breast cancer in histopathology images
Feb 19, 2017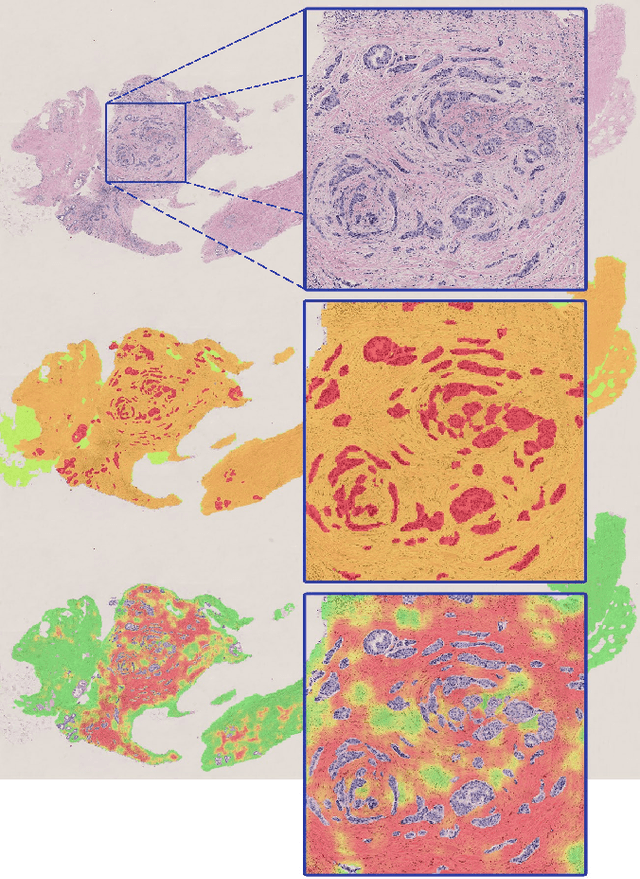
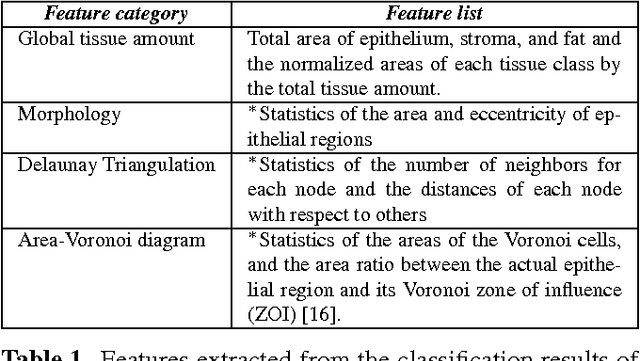
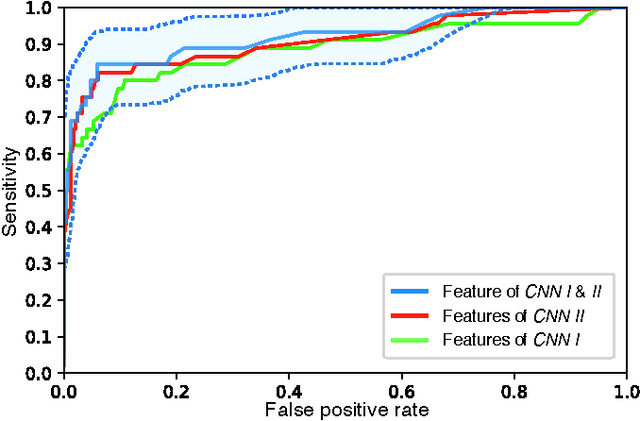
Abstract:Diagnosis of breast carcinomas has so far been limited to the morphological interpretation of epithelial cells and the assessment of epithelial tissue architecture. Consequently, most of the automated systems have focused on characterizing the epithelial regions of the breast to detect cancer. In this paper, we propose a system for classification of hematoxylin and eosin (H&E) stained breast specimens based on convolutional neural networks that primarily targets the assessment of tumor-associated stroma to diagnose breast cancer patients. We evaluate the performance of our proposed system using a large cohort containing 646 breast tissue biopsies. Our evaluations show that the proposed system achieves an area under ROC of 0.92, demonstrating the discriminative power of previously neglected tumor-associated stroma as a diagnostic biomarker.
Location Sensitive Deep Convolutional Neural Networks for Segmentation of White Matter Hyperintensities
Oct 29, 2016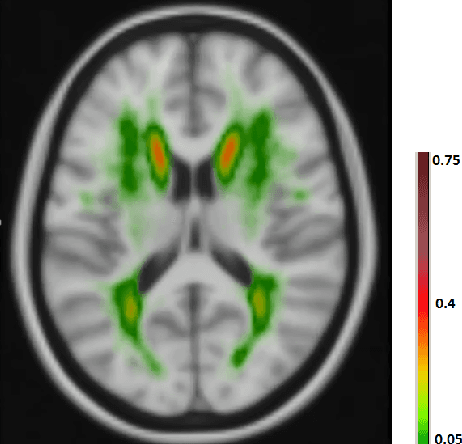

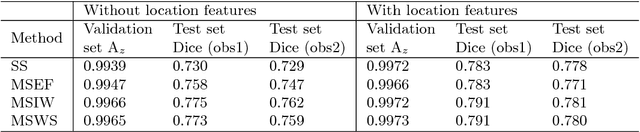
Abstract:The anatomical location of imaging features is of crucial importance for accurate diagnosis in many medical tasks. Convolutional neural networks (CNN) have had huge successes in computer vision, but they lack the natural ability to incorporate the anatomical location in their decision making process, hindering success in some medical image analysis tasks. In this paper, to integrate the anatomical location information into the network, we propose several deep CNN architectures that consider multi-scale patches or take explicit location features while training. We apply and compare the proposed architectures for segmentation of white matter hyperintensities in brain MR images on a large dataset. As a result, we observe that the CNNs that incorporate location information substantially outperform a conventional segmentation method with hand-crafted features as well as CNNs that do not integrate location information. On a test set of 46 scans, the best configuration of our networks obtained a Dice score of 0.791, compared to 0.797 for an independent human observer. Performance levels of the machine and the independent human observer were not statistically significantly different (p-value=0.17).
Deep Multi-scale Location-aware 3D Convolutional Neural Networks for Automated Detection of Lacunes of Presumed Vascular Origin
Oct 29, 2016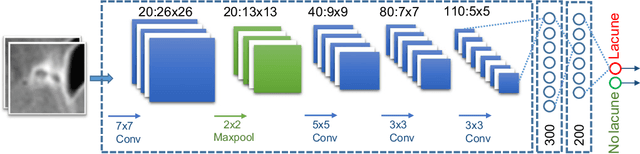
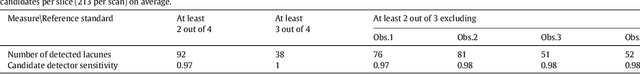
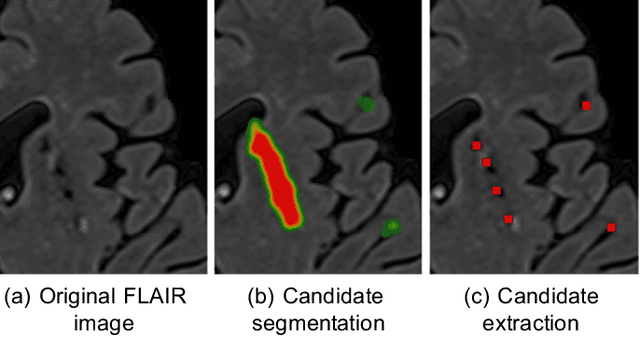
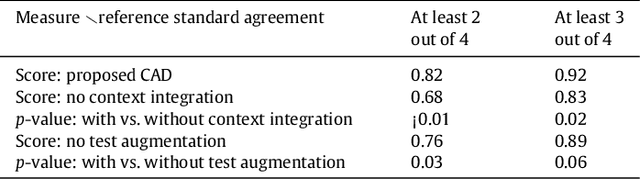
Abstract:Lacunes of presumed vascular origin (lacunes) are associated with an increased risk of stroke, gait impairment, and dementia and are a primary imaging feature of the small vessel disease. Quantification of lacunes may be of great importance to elucidate the mechanisms behind neuro-degenerative disorders and is recommended as part of study standards for small vessel disease research. However, due to the different appearance of lacunes in various brain regions and the existence of other similar-looking structures, such as perivascular spaces, manual annotation is a difficult, elaborative and subjective task, which can potentially be greatly improved by reliable and consistent computer-aided detection (CAD) routines. In this paper, we propose an automated two-stage method using deep convolutional neural networks (CNN). We show that this method has good performance and can considerably benefit readers. We first use a fully convolutional neural network to detect initial candidates. In the second step, we employ a 3D CNN as a false positive reduction tool. As the location information is important to the analysis of candidate structures, we further equip the network with contextual information using multi-scale analysis and integration of explicit location features. We trained, validated and tested our networks on a large dataset of 1075 cases obtained from two different studies. Subsequently, we conducted an observer study with four trained observers and compared our method with them using a free-response operating characteristic analysis. Shown on a test set of 111 cases, the resulting CAD system exhibits performance similar to the trained human observers and achieves a sensitivity of 0.974 with 0.13 false positives per slice. A feasibility study also showed that a trained human observer would considerably benefit once aided by the CAD system.
* 11 pages, 7 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge