Minbyul Jeong
User-Oriented Multi-Turn Dialogue Generation with Tool Use at scale
Jan 13, 2026Abstract:The recent paradigm shift toward large reasoning models (LRMs) as autonomous agents has intensified the demand for sophisticated, multi-turn tool-use capabilities. Yet, existing datasets and data-generation approaches are limited by static, predefined toolsets that cannot scale to the complexity of open-ended human-agent collaboration. To address this, we initially developed a framework for automated task-oriented multi-turn dialogue generation at scale, utilizing an LRM-based simulator to dynamically generate high-value, domain-specific tools to solve specified tasks. However, we observe that a purely task-oriented design often results in "solely task-solving" trajectories, where the agent completes the objective with minimal interaction, failing to generate the high turn-count conversations seen in realistic scenarios. To bridge this gap, we shift toward a user-oriented simulation paradigm. By decoupling task generation from a dedicated user simulator that mimics human behavioral rules - such as incremental request-making and turn-by-turn feedback - we facilitate more authentic, extended multi-turn dialogues that reflect the iterative nature of real-world problem solving. Our generation pipeline operates as a versatile, plug-and-play module capable of initiating generation from any state, ensuring high scalability in producing extended tool-use data. Furthermore, by facilitating multiple task completions within a single trajectory, it yields a high-density dataset that reflects the multifaceted demands of real-world human-agent interaction.
Solar Open Technical Report
Jan 11, 2026Abstract:We introduce Solar Open, a 102B-parameter bilingual Mixture-of-Experts language model for underserved languages. Solar Open demonstrates a systematic methodology for building competitive LLMs by addressing three interconnected challenges. First, to train effectively despite data scarcity for underserved languages, we synthesize 4.5T tokens of high-quality, domain-specific, and RL-oriented data. Second, we coordinate this data through a progressive curriculum jointly optimizing composition, quality thresholds, and domain coverage across 20 trillion tokens. Third, to enable reasoning capabilities through scalable RL, we apply our proposed framework SnapPO for efficient optimization. Across benchmarks in English and Korean, Solar Open achieves competitive performance, demonstrating the effectiveness of this methodology for underserved language AI development.
Trustworthy Agents for Electronic Health Records through Confidence Estimation
Aug 26, 2025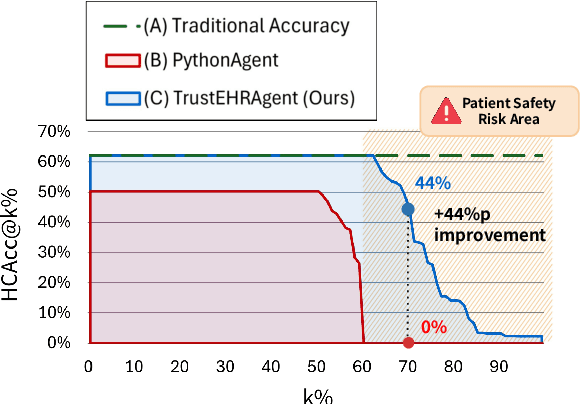
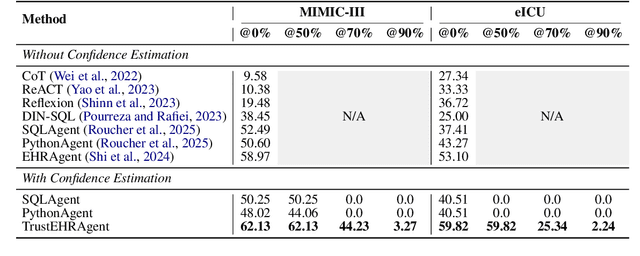
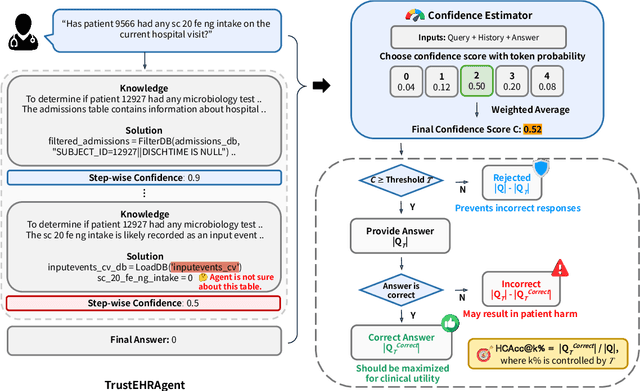
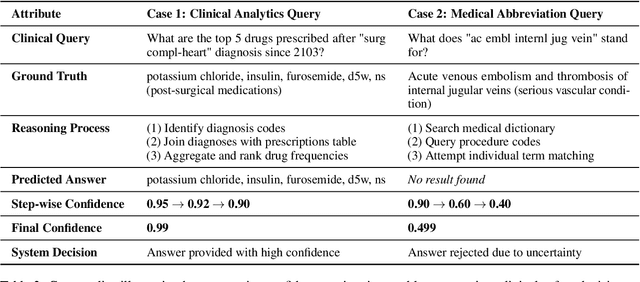
Abstract:Large language models (LLMs) show promise for extracting information from Electronic Health Records (EHR) and supporting clinical decisions. However, deployment in clinical settings faces challenges due to hallucination risks. We propose Hallucination Controlled Accuracy at k% (HCAcc@k%), a novel metric quantifying the accuracy-reliability trade-off at varying confidence thresholds. We introduce TrustEHRAgent, a confidence-aware agent incorporating stepwise confidence estimation for clinical question answering. Experiments on MIMIC-III and eICU datasets show TrustEHRAgent outperforms baselines under strict reliability constraints, achieving improvements of 44.23%p and 25.34%p at HCAcc@70% while baseline methods fail at these thresholds. These results highlight limitations of traditional accuracy metrics in evaluating healthcare AI agents. Our work contributes to developing trustworthy clinical agents that deliver accurate information or transparently express uncertainty when confidence is low.
Does Time Have Its Place? Temporal Heads: Where Language Models Recall Time-specific Information
Feb 20, 2025



Abstract:While the ability of language models to elicit facts has been widely investigated, how they handle temporally changing facts remains underexplored. We discover Temporal Heads, specific attention heads primarily responsible for processing temporal knowledge through circuit analysis. We confirm that these heads are present across multiple models, though their specific locations may vary, and their responses differ depending on the type of knowledge and its corresponding years. Disabling these heads degrades the model's ability to recall time-specific knowledge while maintaining its general capabilities without compromising time-invariant and question-answering performances. Moreover, the heads are activated not only numeric conditions ("In 2004") but also textual aliases ("In the year ..."), indicating that they encode a temporal dimension beyond simple numerical representation. Furthermore, we expand the potential of our findings by demonstrating how temporal knowledge can be edited by adjusting the values of these heads.
ChroKnowledge: Unveiling Chronological Knowledge of Language Models in Multiple Domains
Oct 13, 2024



Abstract:Large language models (LLMs) have significantly impacted many aspects of our lives. However, assessing and ensuring their chronological knowledge remains challenging. Existing approaches fall short in addressing the accumulative nature of knowledge, often relying on a single time stamp. To overcome this, we introduce ChroKnowBench, a benchmark dataset designed to evaluate chronologically accumulated knowledge across three key aspects: multiple domains, time dependency, temporal state. Our benchmark distinguishes between knowledge that evolves (e.g., scientific discoveries, amended laws) and knowledge that remain constant (e.g., mathematical truths, commonsense facts). Building on this benchmark, we present ChroKnowledge (Chronological Categorization of Knowledge), a novel sampling-based framework for evaluating and updating LLMs' non-parametric chronological knowledge. Our evaluation shows: (1) The ability of eliciting temporal knowledge varies depending on the data format that model was trained on. (2) LLMs partially recall knowledge or show a cut-off at temporal boundaries rather than recalling all aspects of knowledge correctly. Thus, we apply our ChroKnowPrompt, an in-depth prompting to elicit chronological knowledge by traversing step-by-step through the surrounding time spans. We observe that our framework successfully updates the overall knowledge across the entire timeline in both the biomedical domain (+11.9%) and the general domain (+2.8%), demonstrating its effectiveness in refining temporal knowledge. This non-parametric approach also enables knowledge updates not only in open-source models but also in proprietary LLMs, ensuring comprehensive applicability across model types. We perform a comprehensive analysis based on temporal characteristics of ChroKnowPrompt and validate the potential of various models to elicit intrinsic temporal knowledge through our method.
CompAct: Compressing Retrieved Documents Actively for Question Answering
Jul 12, 2024Abstract:Retrieval-augmented generation supports language models to strengthen their factual groundings by providing external contexts. However, language models often face challenges when given extensive information, diminishing their effectiveness in solving questions. Context compression tackles this issue by filtering out irrelevant information, but current methods still struggle in realistic scenarios where crucial information cannot be captured with a single-step approach. To overcome this limitation, we introduce CompAct, a novel framework that employs an active strategy to condense extensive documents without losing key information. Our experiments demonstrate that CompAct brings significant improvements in both performance and compression rate on multi-hop question-answering (QA) benchmarks. CompAct flexibly operates as a cost-efficient plug-in module with various off-the-shelf retrievers or readers, achieving exceptionally high compression rates (47x).
OLAPH: Improving Factuality in Biomedical Long-form Question Answering
May 21, 2024



Abstract:In the medical domain, numerous scenarios necessitate the long-form generation ability of large language models (LLMs). Specifically, when addressing patients' questions, it is essential that the model's response conveys factual claims, highlighting the need for an automated method to evaluate those claims. Thus, we introduce MedLFQA, a benchmark dataset reconstructed using long-form question-answering datasets related to the biomedical domain. We use MedLFQA to facilitate the automatic evaluations of factuality. We also propose OLAPH, a simple and novel framework that enables the improvement of factuality through automatic evaluations. The OLAPH framework iteratively trains LLMs to mitigate hallucinations using sampling predictions and preference optimization. In other words, we iteratively set the highest-scoring response as a preferred response derived from sampling predictions and train LLMs to align with the preferred response that improves factuality. We highlight that, even on evaluation metrics not used during training, LLMs trained with our OLAPH framework demonstrate significant performance improvement in factuality. Our findings reveal that a 7B LLM trained with our OLAPH framework can provide long answers comparable to the medical experts' answers in terms of factuality. We believe that our work could shed light on gauging the long-text generation ability of LLMs in the medical domain. Our code and datasets are available at https://github.com/dmis-lab/OLAPH}{https://github.com/dmis-lab/OLAPH.
Improving Medical Reasoning through Retrieval and Self-Reflection with Retrieval-Augmented Large Language Models
Jan 27, 2024



Abstract:Recent proprietary large language models (LLMs), such as GPT-4, have achieved a milestone in tackling diverse challenges in the biomedical domain, ranging from multiple-choice questions to long-form generations. To address challenges that still cannot be handled with the encoded knowledge of LLMs, various retrieval-augmented generation (RAG) methods have been developed by searching documents from the knowledge corpus and appending them unconditionally or selectively to the input of LLMs for generation. However, when applying existing methods to different domain-specific problems, poor generalization becomes apparent, leading to fetching incorrect documents or making inaccurate judgments. In this paper, we introduce Self-BioRAG, a framework reliable for biomedical text that specializes in generating explanations, retrieving domain-specific documents, and self-reflecting generated responses. We utilize 84k filtered biomedical instruction sets to train Self-BioRAG that can assess its generated explanations with customized reflective tokens. Our work proves that domain-specific components, such as a retriever, domain-related document corpus, and instruction sets are necessary for adhering to domain-related instructions. Using three major medical question-answering benchmark datasets, experimental results of Self-BioRAG demonstrate significant performance gains by achieving a 7.2% absolute improvement on average over the state-of-the-art open-foundation model with a parameter size of 7B or less. Overall, we analyze that Self-BioRAG finds the clues in the question, retrieves relevant documents if needed, and understands how to answer with information from retrieved documents and encoded knowledge as a medical expert does. We release our data and code for training our framework components and model weights (7B and 13B) to enhance capabilities in biomedical and clinical domains.
Enhancing Label Consistency on Document-level Named Entity Recognition
Oct 24, 2022Abstract:Named entity recognition (NER) is a fundamental part of extracting information from documents in biomedical applications. A notable advantage of NER is its consistency in extracting biomedical entities in a document context. Although existing document NER models show consistent predictions, they still do not meet our expectations. We investigated whether the adjectives and prepositions within an entity cause a low label consistency, which results in inconsistent predictions. In this paper, we present our method, ConNER, which enhances the label dependency of modifiers (e.g., adjectives and prepositions) to achieve higher label agreement. ConNER refines the draft labels of the modifiers to improve the output representations of biomedical entities. The effectiveness of our method is demonstrated on four popular biomedical NER datasets; in particular, its efficacy is proved on two datasets with 7.5-8.6% absolute improvements in the F1 score. We interpret that our ConNER method is effective on datasets that have intrinsically low label consistency. In the qualitative analysis, we demonstrate how our approach makes the NER model generate consistent predictions. Our code and resources are available at https://github.com/dmis-lab/ConNER/.
BERN2: an advanced neural biomedical named entity recognition and normalization tool
Jan 10, 2022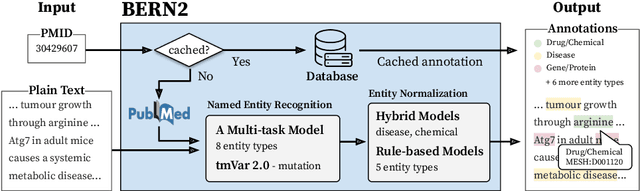
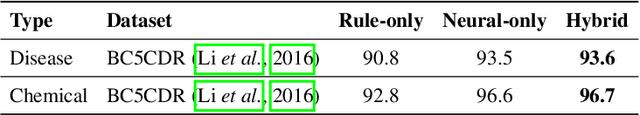
Abstract:In biomedical natural language processing, named entity recognition (NER) and named entity normalization (NEN) are key tasks that enable the automatic extraction of biomedical entities (e.g., diseases and chemicals) from the ever-growing biomedical literature. In this paper, we present BERN2 (Advanced Biomedical Entity Recognition and Normalization), a tool that improves the previous neural network-based NER tool (Kim et al., 2019) by employing a multi-task NER model and neural network-based NEN models to achieve much faster and more accurate inference. We hope that our tool can help annotate large-scale biomedical texts more accurately for various tasks such as biomedical knowledge graph construction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge