Milad Moradi
Artificial intelligence for context-aware visual change detection in software test automation
May 01, 2024Abstract:Automated software testing is integral to the software development process, streamlining workflows and ensuring product reliability. Visual testing within this context, especially concerning user interface (UI) and user experience (UX) validation, stands as one of crucial determinants of overall software quality. Nevertheless, conventional methods like pixel-wise comparison and region-based visual change detection fall short in capturing contextual similarities, nuanced alterations, and understanding the spatial relationships between UI elements. In this paper, we introduce a novel graph-based method for visual change detection in software test automation. Leveraging a machine learning model, our method accurately identifies UI controls from software screenshots and constructs a graph representing contextual and spatial relationships between the controls. This information is then used to find correspondence between UI controls within screenshots of different versions of a software. The resulting graph encapsulates the intricate layout of the UI and underlying contextual relations, providing a holistic and context-aware model. This model is finally used to detect and highlight visual regressions in the UI. Comprehensive experiments on different datasets showed that our change detector can accurately detect visual software changes in various simple and complex test scenarios. Moreover, it outperformed pixel-wise comparison and region-based baselines by a large margin in more complex testing scenarios. This work not only contributes to the advancement of visual change detection but also holds practical implications, offering a robust solution for real-world software test automation challenges, enhancing reliability, and ensuring the seamless evolution of software interfaces.
Multi-level biomedical NER through multi-granularity embeddings and enhanced labeling
Dec 24, 2023Abstract:Biomedical Named Entity Recognition (NER) is a fundamental task of Biomedical Natural Language Processing for extracting relevant information from biomedical texts, such as clinical records, scientific publications, and electronic health records. The conventional approaches for biomedical NER mainly use traditional machine learning techniques, such as Conditional Random Fields and Support Vector Machines or deep learning-based models like Recurrent Neural Networks and Convolutional Neural Networks. Recently, Transformer-based models, including BERT, have been used in the domain of biomedical NER and have demonstrated remarkable results. However, these models are often based on word-level embeddings, limiting their ability to capture character-level information, which is effective in biomedical NER due to the high variability and complexity of biomedical texts. To address these limitations, this paper proposes a hybrid approach that integrates the strengths of multiple models. In this paper, we proposed an approach that leverages fine-tuned BERT to provide contextualized word embeddings, a pre-trained multi-channel CNN for character-level information capture, and following by a BiLSTM + CRF for sequence labelling and modelling dependencies between the words in the text. In addition, also we propose an enhanced labelling method as part of pre-processing to enhance the identification of the entity's beginning word and thus improve the identification of multi-word entities, a common challenge in biomedical NER. By integrating these models and the pre-processing method, our proposed model effectively captures both contextual information and detailed character-level information. We evaluated our model on the benchmark i2b2/2010 dataset, achieving an F1-score of 90.11. These results illustrate the proficiency of our proposed model in performing biomedical Named Entity Recognition.
Model-agnostic explainable artificial intelligence for object detection in image data
Apr 12, 2023



Abstract:Object detection is a fundamental task in computer vision, which has been greatly progressed through developing large and intricate deep learning models. However, the lack of transparency is a big challenge that may not allow the widespread adoption of these models. Explainable artificial intelligence is a field of research where methods are developed to help users understand the behavior, decision logics, and vulnerabilities of AI-based systems. Black-box explanation refers to explaining decisions of an AI system without having access to its internals. In this paper, we design and implement a black-box explanation method named Black-box Object Detection Explanation by Masking (BODEM) through adopting a new masking approach for AI-based object detection systems. We propose local and distant masking to generate multiple versions of an input image. Local masks are used to disturb pixels within a target object to figure out how the object detector reacts to these changes, while distant masks are used to assess how the detection model's decisions are affected by disturbing pixels outside the object. A saliency map is then created by estimating the importance of pixels through measuring the difference between the detection output before and after masking. Finally, a heatmap is created that visualizes how important pixels within the input image are to the detected objects. The experimentations on various object detection datasets and models showed that BODEM can be effectively used to explain the behavior of object detectors and reveal their vulnerabilities. This makes BODEM suitable for explaining and validating AI based object detection systems in black-box software testing scenarios. Furthermore, we conducted data augmentation experiments that showed local masks produced by BODEM can be used for further training the object detectors and improve their detection accuracy and robustness.
ThoughtSource: A central hub for large language model reasoning data
Jan 27, 2023Abstract:Large language models (LLMs) such as GPT-3 and ChatGPT have recently demonstrated impressive results across a wide range of tasks. LLMs are still limited, however, in that they frequently fail at complex reasoning, their reasoning processes are opaque, they are prone to 'hallucinate' facts, and there are concerns about their underlying biases. Letting models verbalize reasoning steps as natural language, a technique known as chain-of-thought prompting, has recently been proposed as a way to address some of these issues. Here we present the first release of ThoughtSource, a meta-dataset and software library for chain-of-thought (CoT) reasoning. The goal of ThoughtSource is to improve future artificial intelligence systems by facilitating qualitative understanding of CoTs, enabling empirical evaluations, and providing training data. This first release of ThoughtSource integrates six scientific/medical, three general-domain and five math word question answering datasets.
A global analysis of metrics used for measuring performance in natural language processing
Apr 25, 2022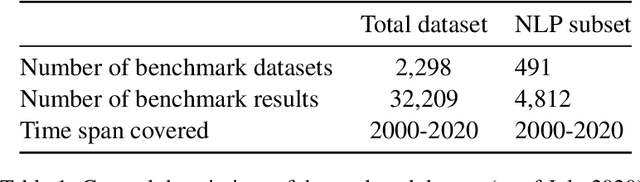
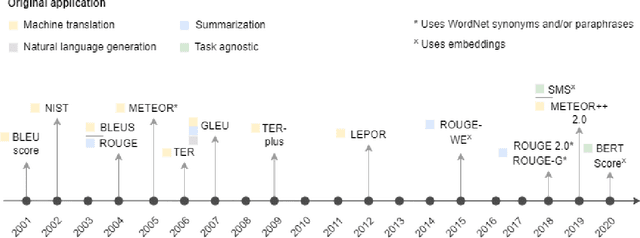
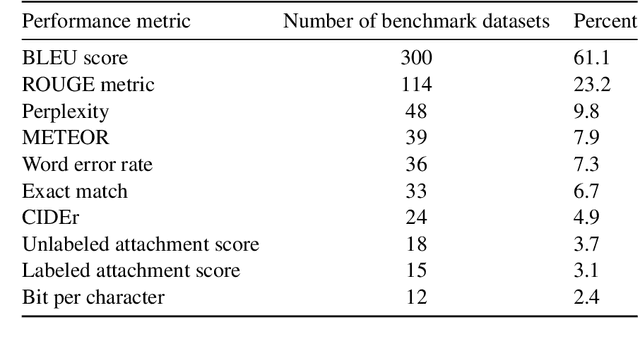
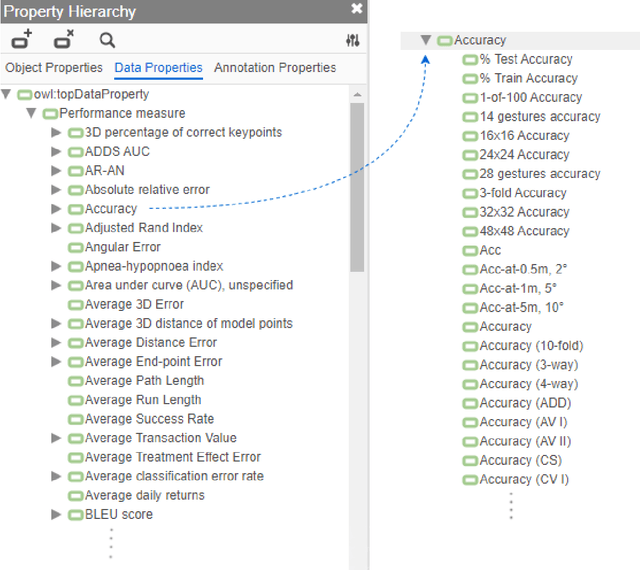
Abstract:Measuring the performance of natural language processing models is challenging. Traditionally used metrics, such as BLEU and ROUGE, originally devised for machine translation and summarization, have been shown to suffer from low correlation with human judgment and a lack of transferability to other tasks and languages. In the past 15 years, a wide range of alternative metrics have been proposed. However, it is unclear to what extent this has had an impact on NLP benchmarking efforts. Here we provide the first large-scale cross-sectional analysis of metrics used for measuring performance in natural language processing. We curated, mapped and systematized more than 3500 machine learning model performance results from the open repository 'Papers with Code' to enable a global and comprehensive analysis. Our results suggest that the large majority of natural language processing metrics currently used have properties that may result in an inadequate reflection of a models' performance. Furthermore, we found that ambiguities and inconsistencies in the reporting of metrics may lead to difficulties in interpreting and comparing model performances, impairing transparency and reproducibility in NLP research.
Deep Learning, Natural Language Processing, and Explainable Artificial Intelligence in the Biomedical Domain
Mar 07, 2022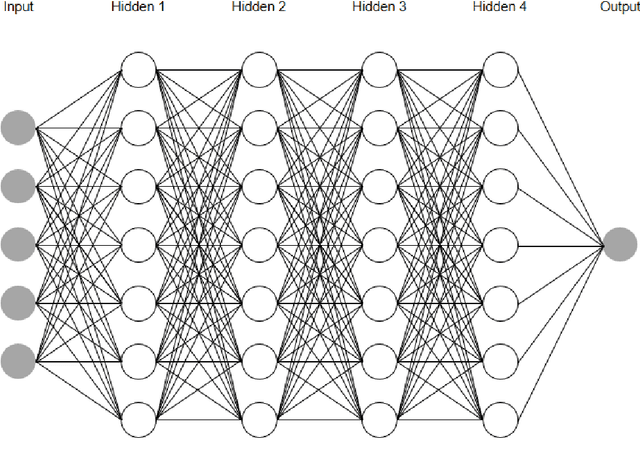
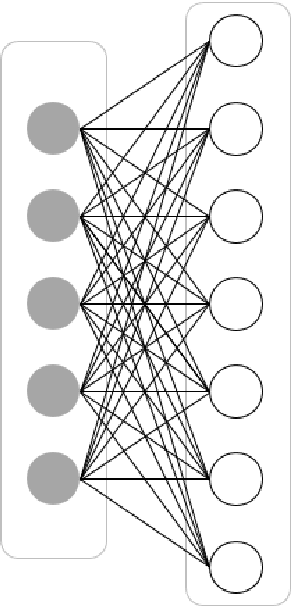
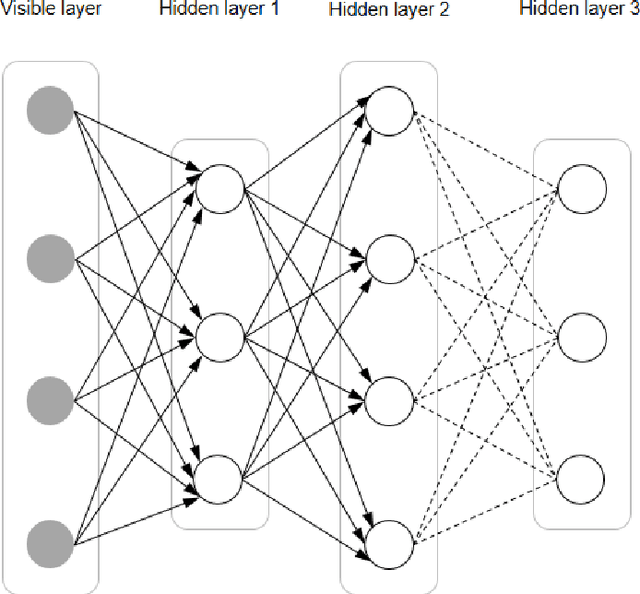
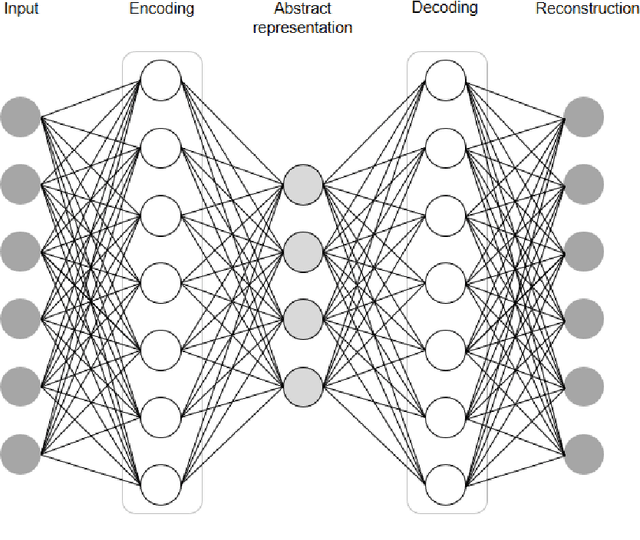
Abstract:In this article, we first give an introduction to artificial intelligence and its applications in biology and medicine in Section 1. Deep learning methods are then described in Section 2. We narrow down the focus of the study on textual data in Section 3, where natural language processing and its applications in the biomedical domain are described. In Section 4, we give an introduction to explainable artificial intelligence and discuss the importance of explainability of artificial intelligence systems, especially in the biomedical domain.
Improving the robustness and accuracy of biomedical language models through adversarial training
Nov 16, 2021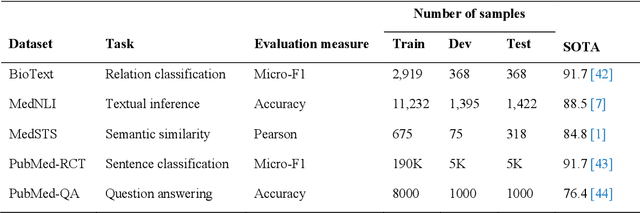
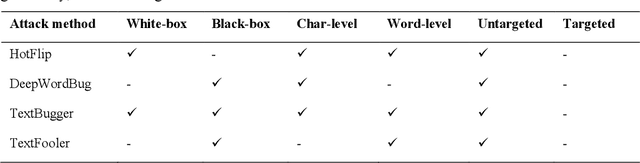
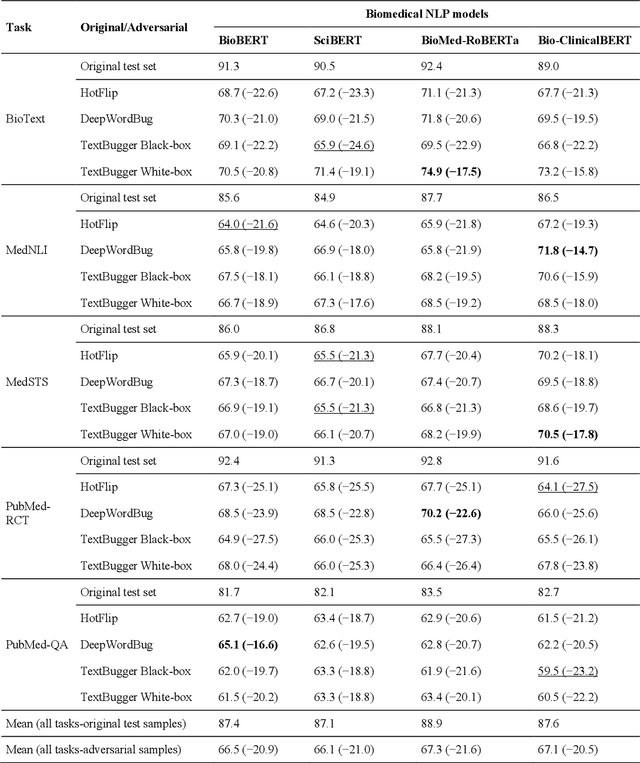
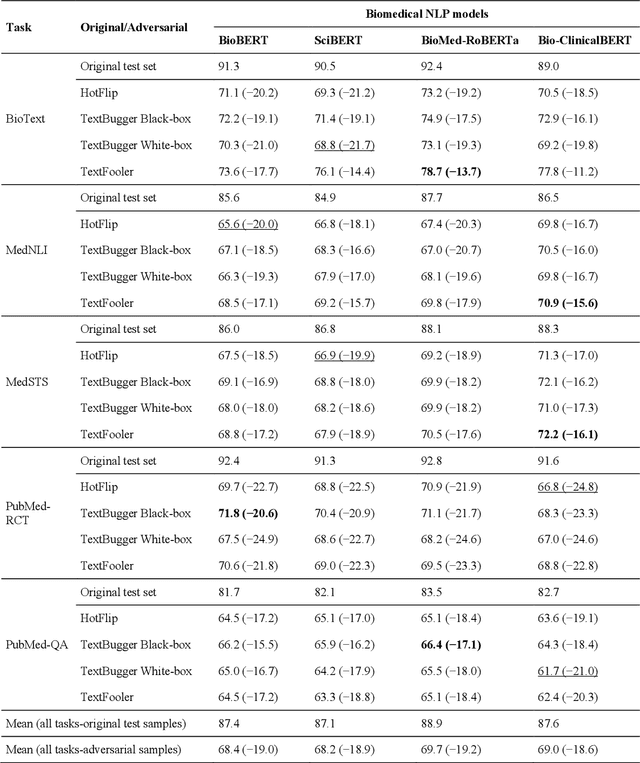
Abstract:Deep transformer neural network models have improved the predictive accuracy of intelligent text processing systems in the biomedical domain. They have obtained state-of-the-art performance scores on a wide variety of biomedical and clinical Natural Language Processing (NLP) benchmarks. However, the robustness and reliability of these models has been less explored so far. Neural NLP models can be easily fooled by adversarial samples, i.e. minor changes to input that preserve the meaning and understandability of the text but force the NLP system to make erroneous decisions. This raises serious concerns about the security and trust-worthiness of biomedical NLP systems, especially when they are intended to be deployed in real-world use cases. We investigated the robustness of several transformer neural language models, i.e. BioBERT, SciBERT, BioMed-RoBERTa, and Bio-ClinicalBERT, on a wide range of biomedical and clinical text processing tasks. We implemented various adversarial attack methods to test the NLP systems in different attack scenarios. Experimental results showed that the biomedical NLP models are sensitive to adversarial samples; their performance dropped in average by 21 and 18.9 absolute percent on character-level and word-level adversarial noise, respectively. Conducting extensive adversarial training experiments, we fine-tuned the NLP models on a mixture of clean samples and adversarial inputs. Results showed that adversarial training is an effective defense mechanism against adversarial noise; the models robustness improved in average by 11.3 absolute percent. In addition, the models performance on clean data increased in average by 2.4 absolute present, demonstrating that adversarial training can boost generalization abilities of biomedical NLP systems.
GPT-3 Models are Poor Few-Shot Learners in the Biomedical Domain
Sep 06, 2021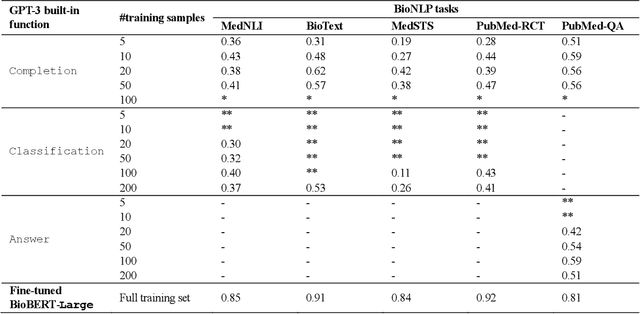
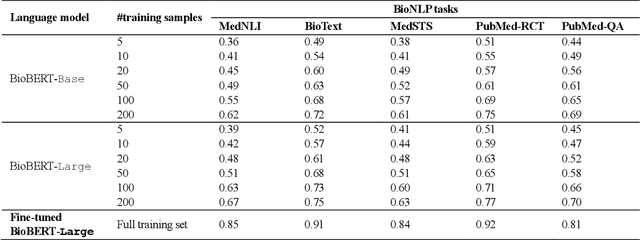
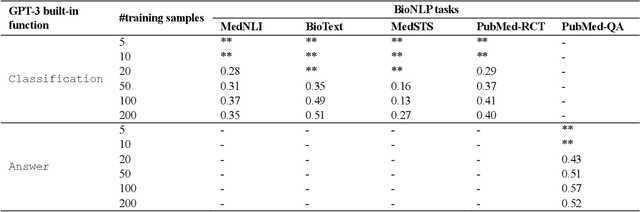
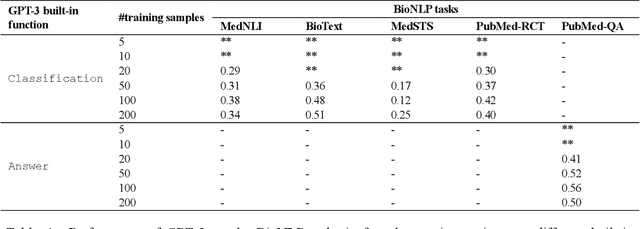
Abstract:Deep neural language models have set new breakthroughs in many tasks of Natural Language Processing (NLP). Recent work has shown that deep transformer language models (pretrained on large amounts of texts) can achieve high levels of task-specific few-shot performance comparable to state-of-the-art models. However, the ability of these large language models in few-shot transfer learning has not yet been explored in the biomedical domain. We investigated the performance of two powerful transformer language models, i.e. GPT-3 and BioBERT, in few-shot settings on various biomedical NLP tasks. The experimental results showed that, to a great extent, both the models underperform a language model fine-tuned on the full training data. Although GPT-3 had already achieved near state-of-the-art results in few-shot knowledge transfer on open-domain NLP tasks, it could not perform as effectively as BioBERT, which is orders of magnitude smaller than GPT-3. Regarding that BioBERT was already pretrained on large biomedical text corpora, our study suggests that language models may largely benefit from in-domain pretraining in task-specific few-shot learning. However, in-domain pretraining seems not to be sufficient; novel pretraining and few-shot learning strategies are required in the biomedical NLP domain.
Deep learning models are not robust against noise in clinical text
Aug 27, 2021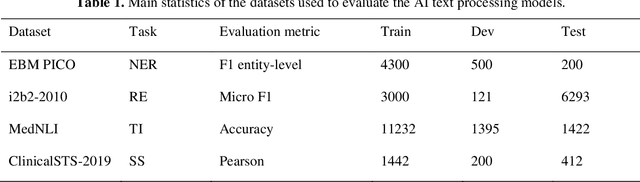
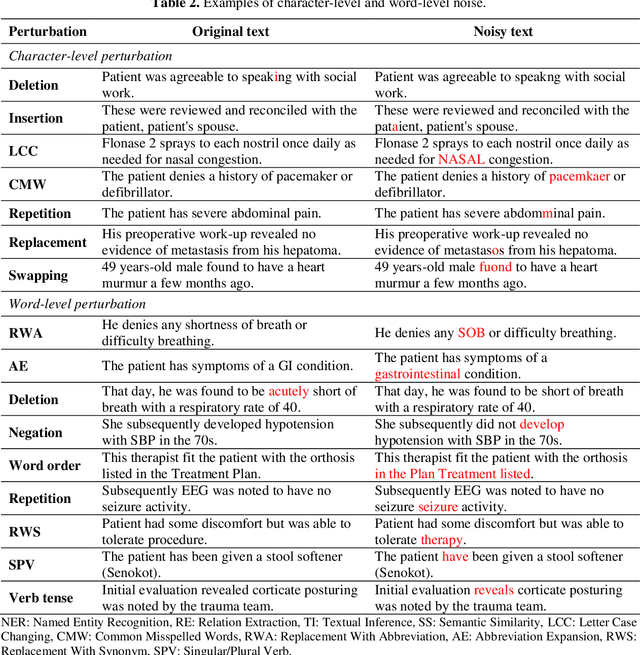
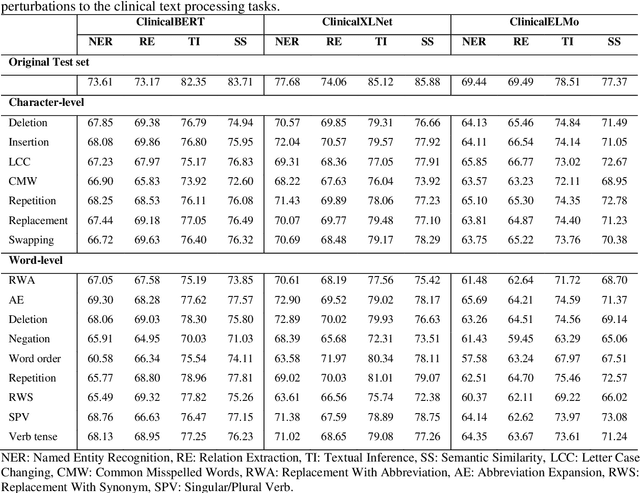
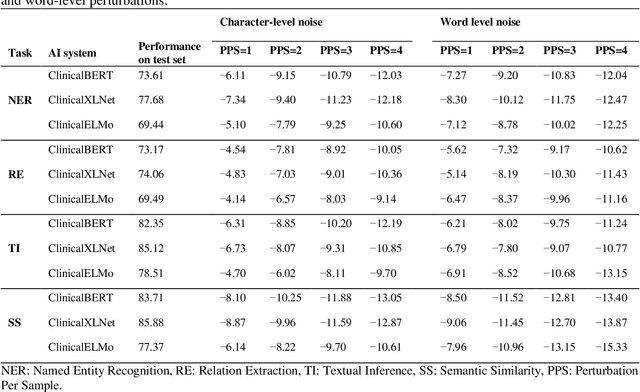
Abstract:Artificial Intelligence (AI) systems are attracting increasing interest in the medical domain due to their ability to learn complicated tasks that require human intelligence and expert knowledge. AI systems that utilize high-performance Natural Language Processing (NLP) models have achieved state-of-the-art results on a wide variety of clinical text processing benchmarks. They have even outperformed human accuracy on some tasks. However, performance evaluation of such AI systems have been limited to accuracy measures on curated and clean benchmark datasets that may not properly reflect how robustly these systems can operate in real-world situations. In order to address this challenge, we introduce and implement a wide variety of perturbation methods that simulate different types of noise and variability in clinical text data. While noisy samples produced by these perturbation methods can often be understood by humans, they may cause AI systems to make erroneous decisions. Conducting extensive experiments on several clinical text processing tasks, we evaluated the robustness of high-performance NLP models against various types of character-level and word-level noise. The results revealed that the NLP models performance degrades when the input contains small amounts of noise. This study is a significant step towards exposing vulnerabilities of AI models utilized in clinical text processing systems. The proposed perturbation methods can be used in performance evaluation tests to assess how robustly clinical NLP models can operate on noisy data, in real-world settings.
Evaluating the Robustness of Neural Language Models to Input Perturbations
Aug 27, 2021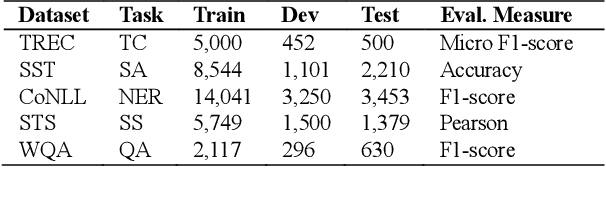
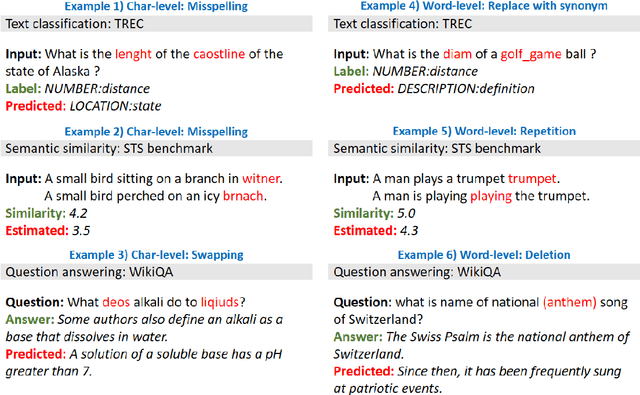
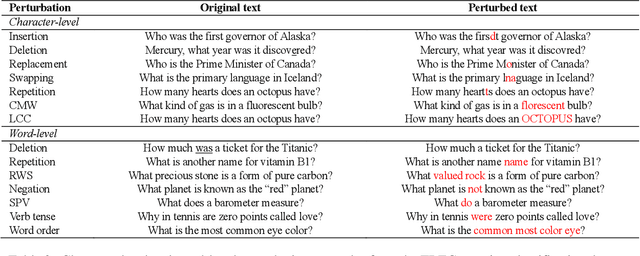
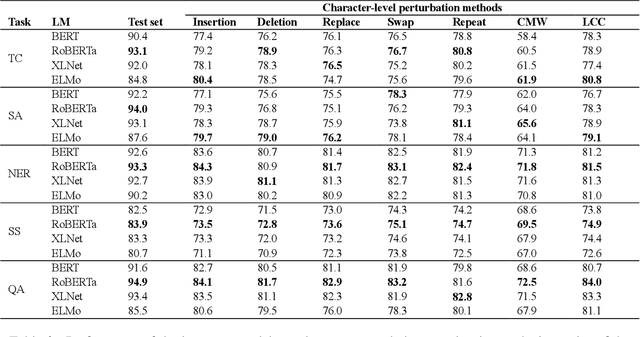
Abstract:High-performance neural language models have obtained state-of-the-art results on a wide range of Natural Language Processing (NLP) tasks. However, results for common benchmark datasets often do not reflect model reliability and robustness when applied to noisy, real-world data. In this study, we design and implement various types of character-level and word-level perturbation methods to simulate realistic scenarios in which input texts may be slightly noisy or different from the data distribution on which NLP systems were trained. Conducting comprehensive experiments on different NLP tasks, we investigate the ability of high-performance language models such as BERT, XLNet, RoBERTa, and ELMo in handling different types of input perturbations. The results suggest that language models are sensitive to input perturbations and their performance can decrease even when small changes are introduced. We highlight that models need to be further improved and that current benchmarks are not reflecting model robustness well. We argue that evaluations on perturbed inputs should routinely complement widely-used benchmarks in order to yield a more realistic understanding of NLP systems robustness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge