Michael Hughes
Brown University
Entropic Analysis of Time Series through Kernel Density Estimation
Mar 24, 2025Abstract:This work presents a novel framework for time series analysis using entropic measures based on the kernel density estimate (KDE) of the time series' Takens' embeddings. Using this framework we introduce two distinct analytical tools: (1) a multi-scale KDE entropy metric, denoted as $\Delta\text{KE}$, which quantifies the evolution of time series complexity across different scales by measuring certain entropy changes, and (2) a sliding baseline method that employs the Kullback-Leibler (KL) divergence to detect changes in time series dynamics through changes in KDEs. The $\Delta{\rm KE}$ metric offers insights into the information content and ``unfolding'' properties of the time series' embedding related to dynamical systems, while the KL divergence-based approach provides a noise and outlier robust approach for identifying time series change points (injections in RF signals, e.g.). We demonstrate the versatility and effectiveness of these tools through a set of experiments encompassing diverse domains. In the space of radio frequency (RF) signal processing, we achieve accurate detection of signal injections under varying noise and interference conditions. Furthermore, we apply our methodology to electrocardiography (ECG) data, successfully identifying instances of ventricular fibrillation with high accuracy. Finally, we demonstrate the potential of our tools for dynamic state detection by accurately identifying chaotic regimes within an intermittent signal. These results show the broad applicability of our framework for extracting meaningful insights from complex time series data across various scientific disciplines.
Regional Tree Regularization for Interpretability in Black Box Models
Aug 13, 2019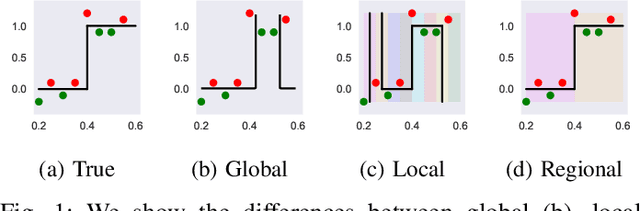
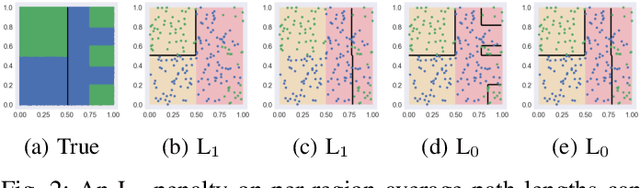
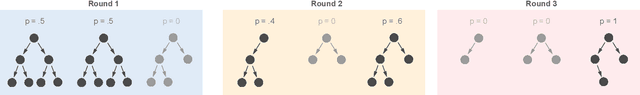
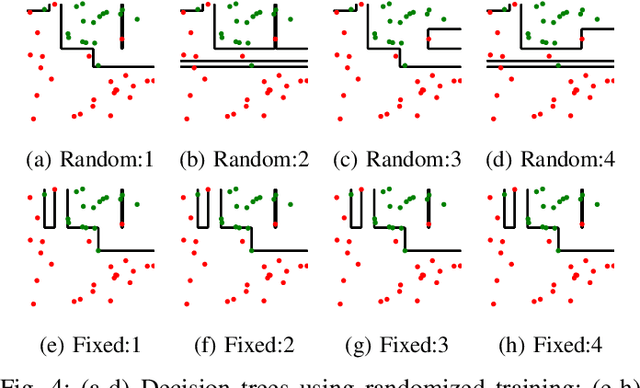
Abstract:The lack of interpretability remains a barrier to the adoption of deep neural networks. Recently, tree regularization has been proposed to encourage deep neural networks to resemble compact, axis-aligned decision trees without significant compromises in accuracy. However, it may be unreasonable to expect that a single tree can predict well across all possible inputs. In this work, we propose regional tree regularization, which encourages a deep model to be well-approximated by several separate decision trees specific to predefined regions of the input space. Practitioners can define regions based on domain knowledge of contexts where different decision-making logic is needed. Across many datasets, our approach delivers more accurate predictions than simply training separate decision trees for each region, while producing simpler explanations than other neural net regularization schemes without sacrificing predictive power. Two healthcare case studies in critical care and HIV demonstrate how experts can improve understanding of deep models via our approach.
Intraoperative robotic-assisted large-area high-speed microscopic imaging and intervention
Aug 13, 2018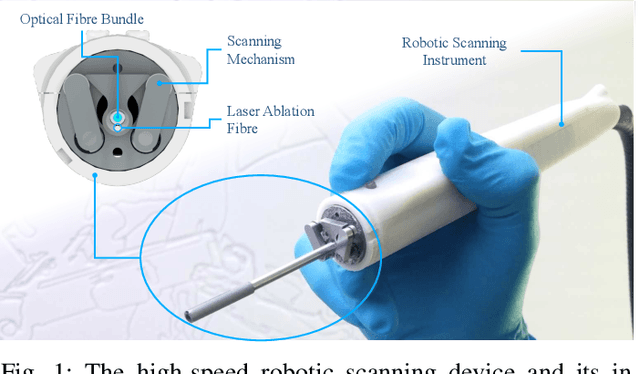
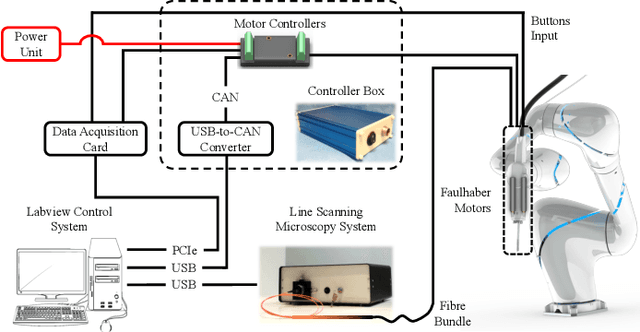
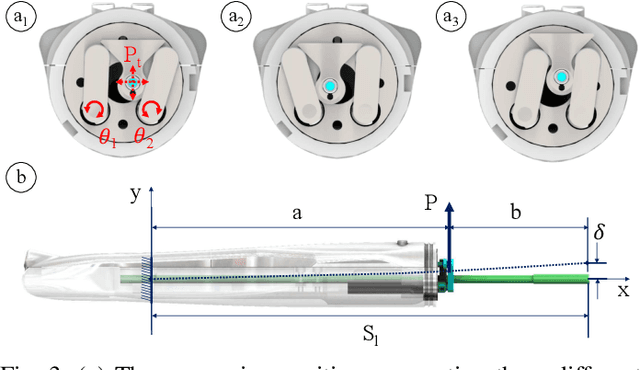
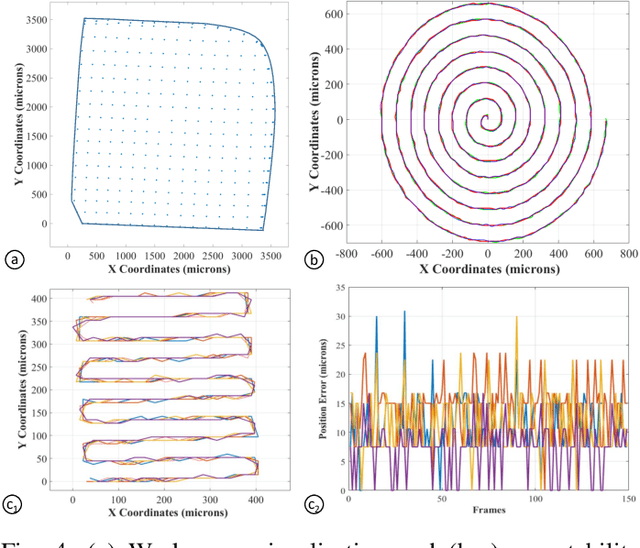
Abstract:Objective: Probe-based confocal endomicroscopy is an emerging high-magnification optical imaging technique that provides in vivo and in situ cellular-level imaging for real-time assessment of tissue pathology. Endomicroscopy could potentially be used for intraoperative surgical guidance, but it is challenging to assess a surgical site using individual microscopic images due to the limited field-of-view and difficulties associated with manually manipulating the probe. Methods: In this paper, a novel robotic device for large-area endomicroscopy imaging is proposed, demonstrating a rapid, but highly accurate, scanning mechanism with image-based motion control which is able to generate histology-like endomicroscopy mosaics. The device also includes, for the first time in robotic-assisted endomicroscopy, the capability to ablate tissue without the need for an additional tool. Results: The device achieves pre-programmed trajectories with positioning accuracy of less than 30 um, while the image-based approach demonstrated that it can suppress random motion disturbances up to 1.25 mm/s. Mosaics are presented from a range of ex vivo human and animal tissues, over areas of more than 3 mm^2, scanned in approximate 10 seconds. Conclusion: This work demonstrates the potential of the proposed instrument to generate large-area, high-resolution microscopic images for intraoperative tissue identification and margin assessment. Significance: This approach presents an important alternative to current histology techniques, significantly reducing the tissue assessment time, while simultaneously providing the capability to mark and ablate suspicious areas intraoperatively.
Autonomous Scanning for Endomicroscopic Mosaicing and 3D Fusion
Oct 21, 2016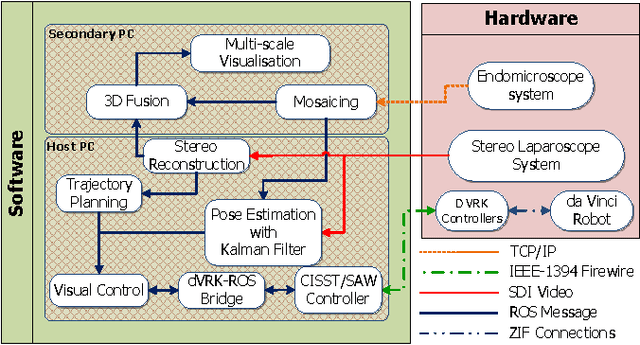
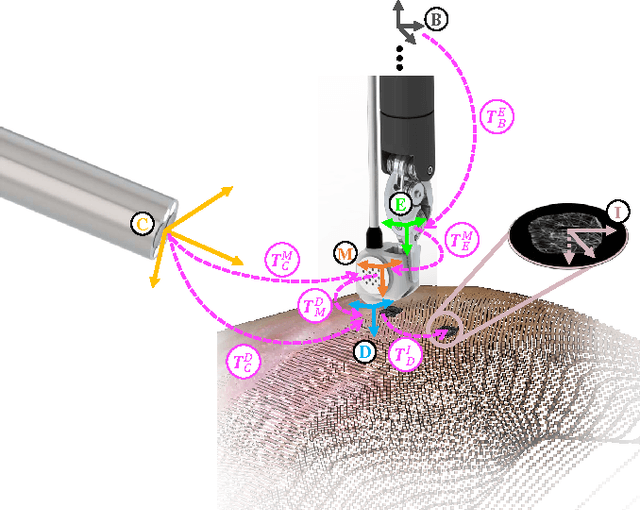
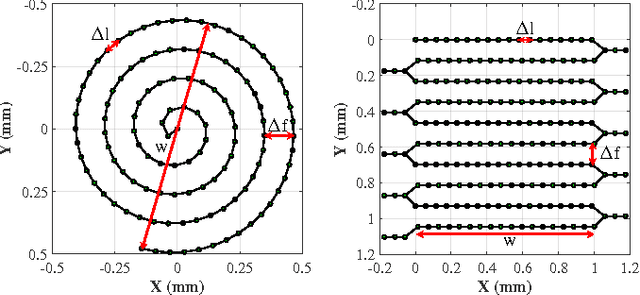
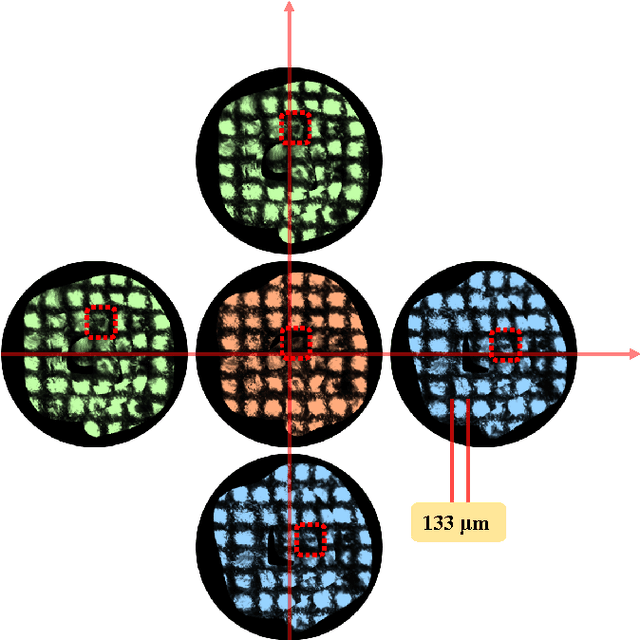
Abstract:Robotic-assisted Minimally Invasive Surgery (RMIS) can benefit from the automation of common, repetitive or well-defined but ergonomically difficult tasks. One such task is the scanning of a pick-up endomicroscopy probe over a complex, undulating tissue surface in order to enhance the effective field-of-view through video mosaicing. In this paper, the da Vinci surgical robot, through the dVRK framework, is used for autonomous scanning and 2D mosaicing over a user-defined region of interest. To achieve the level of precision required for high quality large-area mosaic generation, which relies on sufficient overlap between consecutive image frames, visual servoing is performed using a tracking marker attached to the probe. The resulting sub-millimetre accuracy of the probe motion allows for the generation of large endomicroscopy mo- saics with minimal intervention from the surgeon. It also allows the probe to be maintained in an orientation perpendicular to the local tissue surface, providing optimal imaging results. Images are streamed from the endomicroscope and overlaid live onto the surgeons view, while 2D mosaics are generated in real-time, and fused into a 3D stereo reconstruction of the surgical scene, thus providing intuitive visualisation and fusion of the multi-scale images. The system therefore offers significant potential to enhance surgical procedures, by providing the operator with cellular-scale information over a larger area than could typically be achieved by manual scanning.
The Nonparametric Metadata Dependent Relational Model
Jun 27, 2012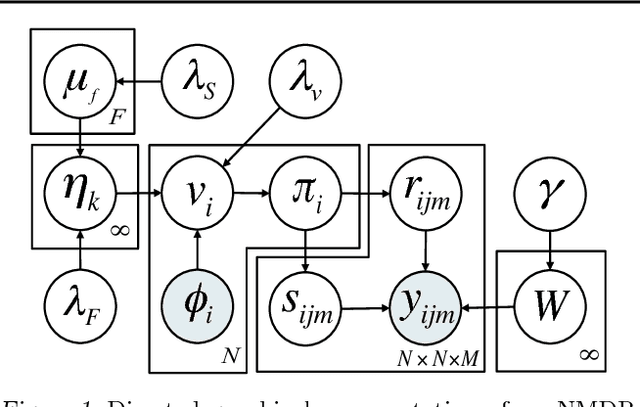
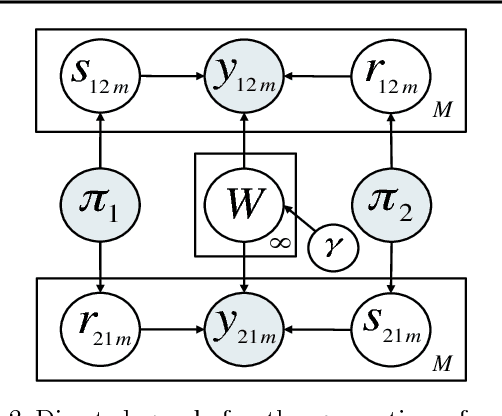
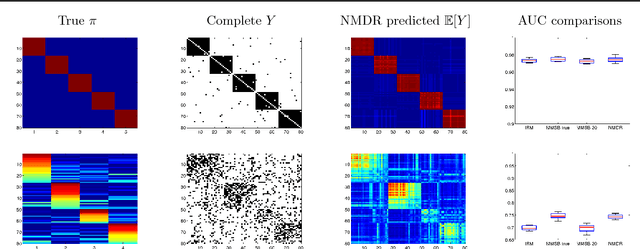
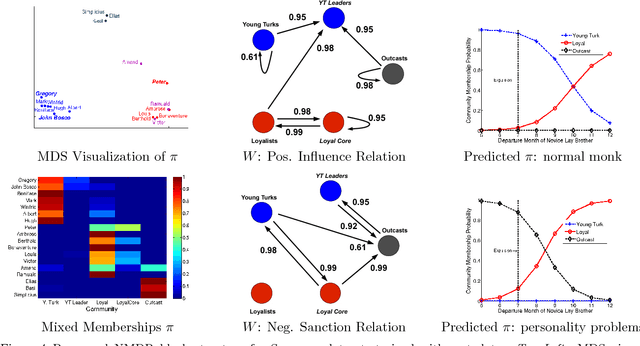
Abstract:We introduce the nonparametric metadata dependent relational (NMDR) model, a Bayesian nonparametric stochastic block model for network data. The NMDR allows the entities associated with each node to have mixed membership in an unbounded collection of latent communities. Learned regression models allow these memberships to depend on, and be predicted from, arbitrary node metadata. We develop efficient MCMC algorithms for learning NMDR models from partially observed node relationships. Retrospective MCMC methods allow our sampler to work directly with the infinite stick-breaking representation of the NMDR, avoiding the need for finite truncations. Our results demonstrate recovery of useful latent communities from real-world social and ecological networks, and the usefulness of metadata in link prediction tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge