Michael A. Rosenberg
Signal, Image, or Symbolic: Exploring the Best Input Representation for Electrocardiogram-Language Models Through a Unified Framework
May 24, 2025Abstract:Recent advances have increasingly applied large language models (LLMs) to electrocardiogram (ECG) interpretation, giving rise to Electrocardiogram-Language Models (ELMs). Conditioned on an ECG and a textual query, an ELM autoregressively generates a free-form textual response. Unlike traditional classification-based systems, ELMs emulate expert cardiac electrophysiologists by issuing diagnoses, analyzing waveform morphology, identifying contributing factors, and proposing patient-specific action plans. To realize this potential, researchers are curating instruction-tuning datasets that pair ECGs with textual dialogues and are training ELMs on these resources. Yet before scaling ELMs further, there is a fundamental question yet to be explored: What is the most effective ECG input representation? In recent works, three candidate representations have emerged-raw time-series signals, rendered images, and discretized symbolic sequences. We present the first comprehensive benchmark of these modalities across 6 public datasets and 5 evaluation metrics. We find symbolic representations achieve the greatest number of statistically significant wins over both signal and image inputs. We further ablate the LLM backbone, ECG duration, and token budget, and we evaluate robustness to signal perturbations. We hope that our findings offer clear guidance for selecting input representations when developing the next generation of ELMs.
ECG-Byte: A Tokenizer for End-to-End Generative Electrocardiogram Language Modeling
Dec 18, 2024



Abstract:Large Language Models (LLMs) have shown remarkable adaptability across domains beyond text, specifically electrocardiograms (ECGs). More specifically, there is a growing body of work exploring the task of generating text from a multi-channeled ECG and corresponding textual prompt. Current approaches typically involve pretraining an ECG-specific encoder with a self-supervised learning (SSL) objective and using the features output by the pretrained encoder to finetune a LLM for natural language generation (NLG). However, these methods are limited by 1) inefficiency from two-stage training and 2) interpretability challenges with encoder-generated features. To address these limitations, we introduce ECG-Byte, an adapted byte pair encoding (BPE) tokenizer pipeline for autoregressive language modeling of ECGs. This approach compresses and encodes ECG signals into tokens, enabling end-to-end LLM training by combining ECG and text tokens directly, while being much more interpretable since the ECG tokens can be directly mapped back to the original signal. Using ECG-Byte, we achieve competitive performance in NLG tasks in only half the time and ~48% of the data required by two-stage approaches.
Interpretation of Intracardiac Electrograms Through Textual Representations
Feb 02, 2024



Abstract:Understanding the irregular electrical activity of atrial fibrillation (AFib) has been a key challenge in electrocardiography. For serious cases of AFib, catheter ablations are performed to collect intracardiac electrograms (EGMs). EGMs offer intricately detailed and localized electrical activity of the heart and are an ideal modality for interpretable cardiac studies. Recent advancements in artificial intelligence (AI) has allowed some works to utilize deep learning frameworks to interpret EGMs during AFib. Additionally, language models (LMs) have shown exceptional performance in being able to generalize to unseen domains, especially in healthcare. In this study, we are the first to leverage pretrained LMs for finetuning of EGM interpolation and AFib classification via masked language modeling. We formulate the EGM as a textual sequence and present competitive performances on AFib classification compared against other representations. Lastly, we provide a comprehensive interpretability study to provide a multi-perspective intuition of the model's behavior, which could greatly benefit the clinical use.
GeoECG: Data Augmentation via Wasserstein Geodesic Perturbation for Robust Electrocardiogram Prediction
Aug 10, 2022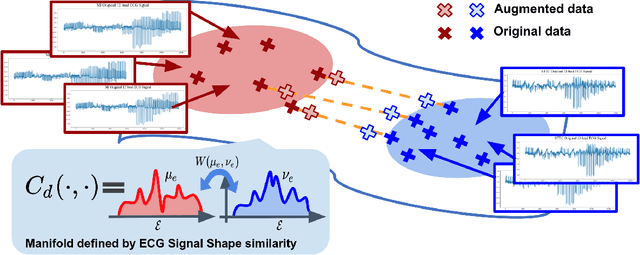
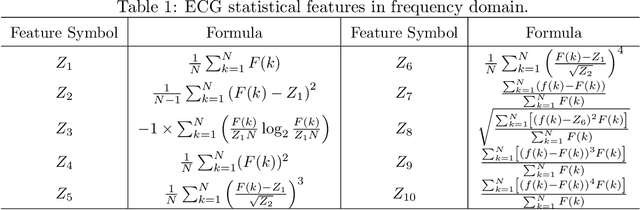

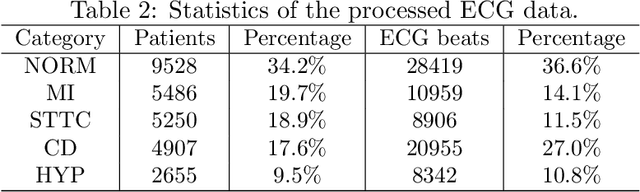
Abstract:There has been an increased interest in applying deep neural networks to automatically interpret and analyze the 12-lead electrocardiogram (ECG). The current paradigms with machine learning methods are often limited by the amount of labeled data. This phenomenon is particularly problematic for clinically-relevant data, where labeling at scale can be time-consuming and costly in terms of the specialized expertise and human effort required. Moreover, deep learning classifiers may be vulnerable to adversarial examples and perturbations, which could have catastrophic consequences, for example, when applied in the context of medical treatment, clinical trials, or insurance claims. In this paper, we propose a physiologically-inspired data augmentation method to improve performance and increase the robustness of heart disease detection based on ECG signals. We obtain augmented samples by perturbing the data distribution towards other classes along the geodesic in Wasserstein space. To better utilize domain-specific knowledge, we design a ground metric that recognizes the difference between ECG signals based on physiologically determined features. Learning from 12-lead ECG signals, our model is able to distinguish five categories of cardiac conditions. Our results demonstrate improvements in accuracy and robustness, reflecting the effectiveness of our data augmentation method.
* 26 pages, Figure 13, Machine Learning for Healthcare 2022
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge