Max Welling
UC Irvine
Pixelated Reconstruction of Foreground Density and Background Surface Brightness in Gravitational Lensing Systems using Recurrent Inference Machines
Jan 10, 2023Abstract:Modeling strong gravitational lenses in order to quantify the distortions in the images of background sources and to reconstruct the mass density in the foreground lenses has been a difficult computational challenge. As the quality of gravitational lens images increases, the task of fully exploiting the information they contain becomes computationally and algorithmically more difficult. In this work, we use a neural network based on the Recurrent Inference Machine (RIM) to simultaneously reconstruct an undistorted image of the background source and the lens mass density distribution as pixelated maps. The method iteratively reconstructs the model parameters (the image of the source and a pixelated density map) by learning the process of optimizing the likelihood given the data using the physical model (a ray-tracing simulation), regularized by a prior implicitly learned by the neural network through its training data. When compared to more traditional parametric models, the proposed method is significantly more expressive and can reconstruct complex mass distributions, which we demonstrate by using realistic lensing galaxies taken from the IllustrisTNG cosmological hydrodynamic simulation.
Structure-based Drug Design with Equivariant Diffusion Models
Oct 24, 2022Abstract:Structure-based drug design (SBDD) aims to design small-molecule ligands that bind with high affinity and specificity to pre-determined protein targets. Traditional SBDD pipelines start with large-scale docking of compound libraries from public databases, thus limiting the exploration of chemical space to existent previously studied regions. Recent machine learning methods approached this problem using an atom-by-atom generation approach, which is computationally expensive. In this paper, we formulate SBDD as a 3D-conditional generation problem and present DiffSBDD, an E(3)-equivariant 3D-conditional diffusion model that generates novel ligands conditioned on protein pockets. Furthermore, we curate a new dataset of experimentally determined binding complex data from Binding MOAD to provide a realistic binding scenario that complements the synthetic CrossDocked dataset. Comprehensive in silico experiments demonstrate the efficiency of DiffSBDD in generating novel and diverse drug-like ligands that engage protein pockets with high binding energies as predicted by in silico docking.
Equivariant 3D-Conditional Diffusion Models for Molecular Linker Design
Oct 11, 2022
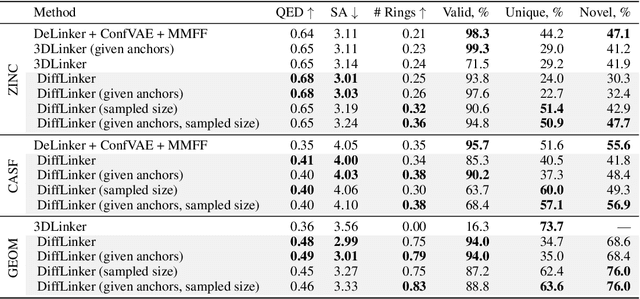
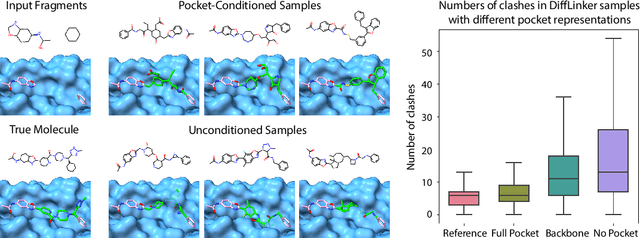
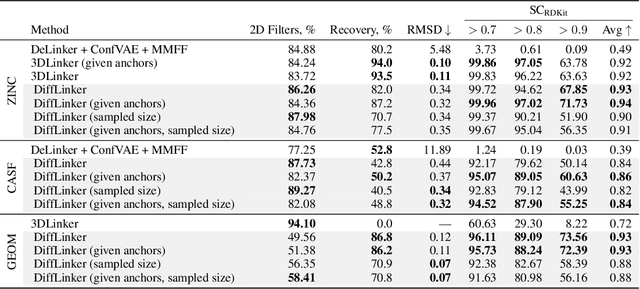
Abstract:Fragment-based drug discovery has been an effective paradigm in early-stage drug development. An open challenge in this area is designing linkers between disconnected molecular fragments of interest to obtain chemically-relevant candidate drug molecules. In this work, we propose DiffLinker, an E(3)-equivariant 3D-conditional diffusion model for molecular linker design. Given a set of disconnected fragments, our model places missing atoms in between and designs a molecule incorporating all the initial fragments. Unlike previous approaches that are only able to connect pairs of molecular fragments, our method can link an arbitrary number of fragments. Additionally, the model automatically determines the number of atoms in the linker and its attachment points to the input fragments. We demonstrate that DiffLinker outperforms other methods on the standard datasets generating more diverse and synthetically-accessible molecules. Besides, we experimentally test our method in real-world applications, showing that it can successfully generate valid linkers conditioned on target protein pockets.
SVNet: Where SO(3) Equivariance Meets Binarization on Point Cloud Representation
Sep 20, 2022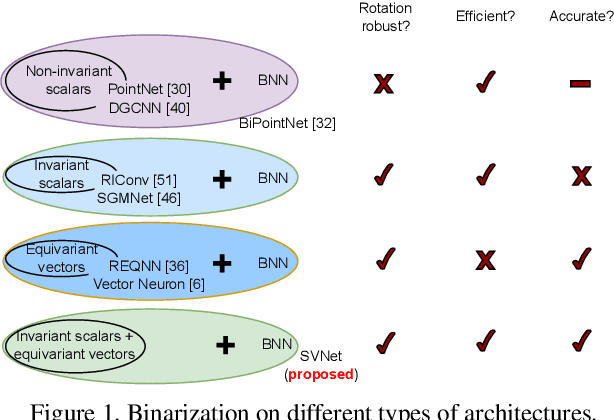
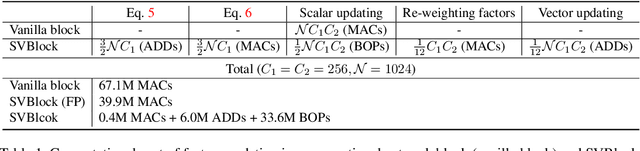
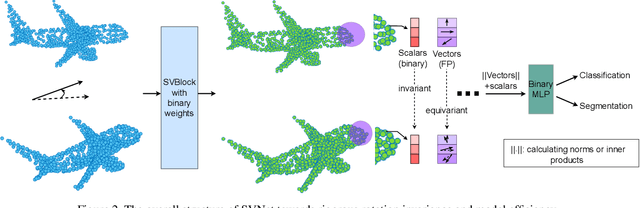
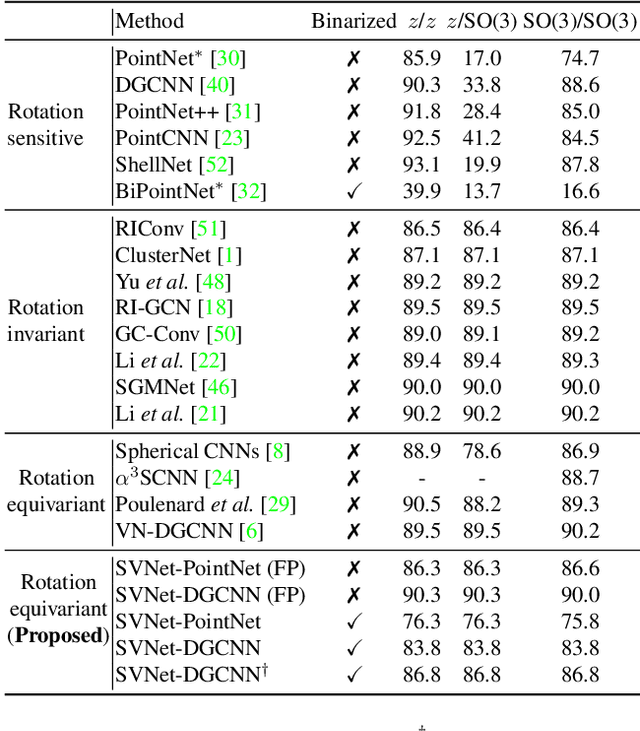
Abstract:Efficiency and robustness are increasingly needed for applications on 3D point clouds, with the ubiquitous use of edge devices in scenarios like autonomous driving and robotics, which often demand real-time and reliable responses. The paper tackles the challenge by designing a general framework to construct 3D learning architectures with SO(3) equivariance and network binarization. However, a naive combination of equivariant networks and binarization either causes sub-optimal computational efficiency or geometric ambiguity. We propose to locate both scalar and vector features in our networks to avoid both cases. Precisely, the presence of scalar features makes the major part of the network binarizable, while vector features serve to retain rich structural information and ensure SO(3) equivariance. The proposed approach can be applied to general backbones like PointNet and DGCNN. Meanwhile, experiments on ModelNet40, ShapeNet, and the real-world dataset ScanObjectNN, demonstrated that the method achieves a great trade-off between efficiency, rotation robustness, and accuracy. The codes are available at https://github.com/zhuoinoulu/svnet.
Clifford Neural Layers for PDE Modeling
Sep 08, 2022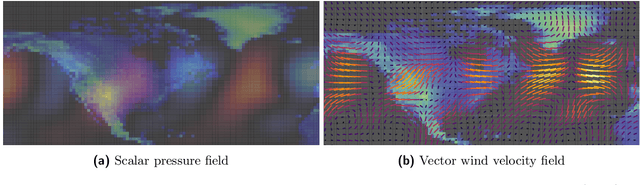
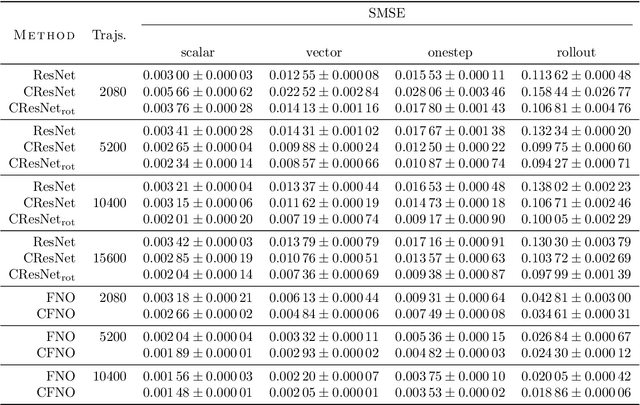
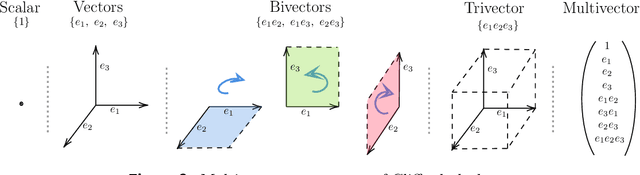
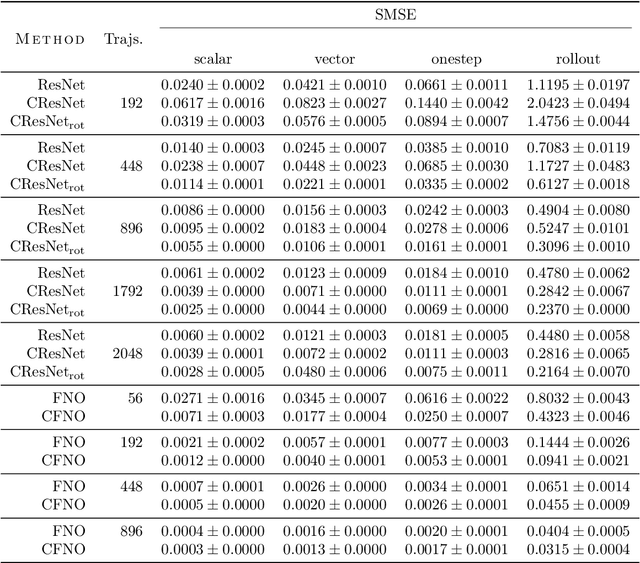
Abstract:Partial differential equations (PDEs) see widespread use in sciences and engineering to describe simulation of physical processes as scalar and vector fields interacting and coevolving over time. Due to the computationally expensive nature of their standard solution methods, neural PDE surrogates have become an active research topic to accelerate these simulations. However, current methods do not explicitly take into account the relationship between different fields and their internal components, which are often correlated. Viewing the time evolution of such correlated fields through the lens of multivector fields allows us to overcome these limitations. Multivector fields consist of scalar, vector, as well as higher-order components, such as bivectors and trivectors. Their algebraic properties, such as multiplication, addition and other arithmetic operations can be described by Clifford algebras. To our knowledge, this paper presents the first usage of such multivector representations together with Clifford convolutions and Clifford Fourier transforms in the context of deep learning. The resulting Clifford neural layers are universally applicable and will find direct use in the areas of fluid dynamics, weather forecasting, and the modeling of physical systems in general. We empirically evaluate the benefit of Clifford neural layers by replacing convolution and Fourier operations in common neural PDE surrogates by their Clifford counterparts on two-dimensional Navier-Stokes and weather modeling tasks, as well as three-dimensional Maxwell equations. Clifford neural layers consistently improve generalization capabilities of the tested neural PDE surrogates.
Bayesian Optimization for Macro Placement
Jul 18, 2022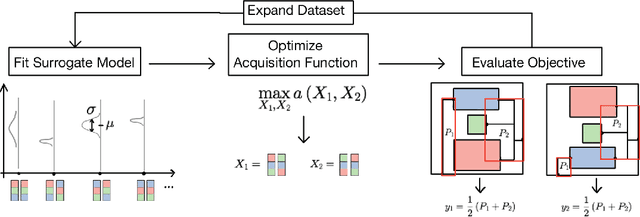
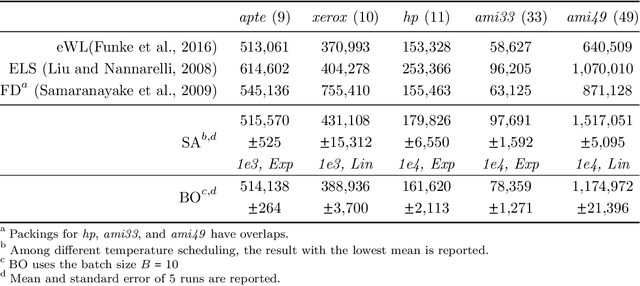
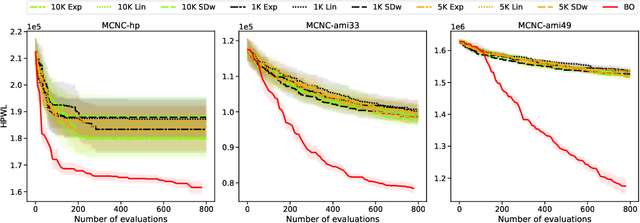
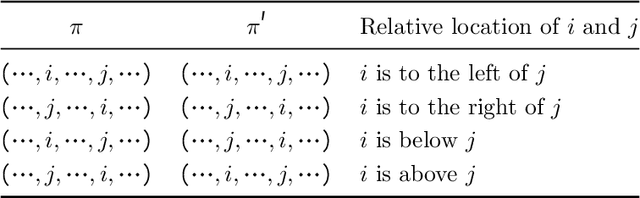
Abstract:Macro placement is the problem of placing memory blocks on a chip canvas. It can be formulated as a combinatorial optimization problem over sequence pairs, a representation which describes the relative positions of macros. Solving this problem is particularly challenging since the objective function is expensive to evaluate. In this paper, we develop a novel approach to macro placement using Bayesian optimization (BO) over sequence pairs. BO is a machine learning technique that uses a probabilistic surrogate model and an acquisition function that balances exploration and exploitation to efficiently optimize a black-box objective function. BO is more sample-efficient than reinforcement learning and therefore can be used with more realistic objectives. Additionally, the ability to learn from data and adapt the algorithm to the objective function makes BO an appealing alternative to other black-box optimization methods such as simulated annealing, which relies on problem-dependent heuristics and parameter-tuning. We benchmark our algorithm on the fixed-outline macro placement problem with the half-perimeter wire length objective and demonstrate competitive performance.
Path Integral Stochastic Optimal Control for Sampling Transition Paths
Jun 27, 2022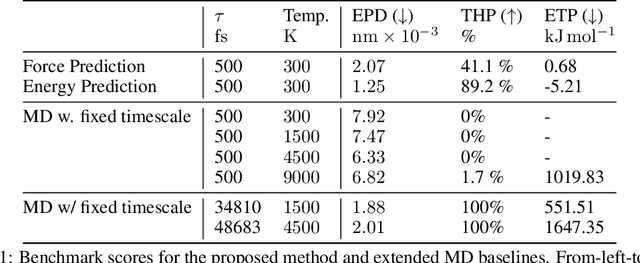
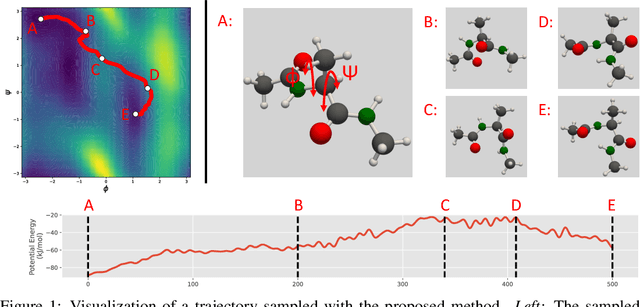
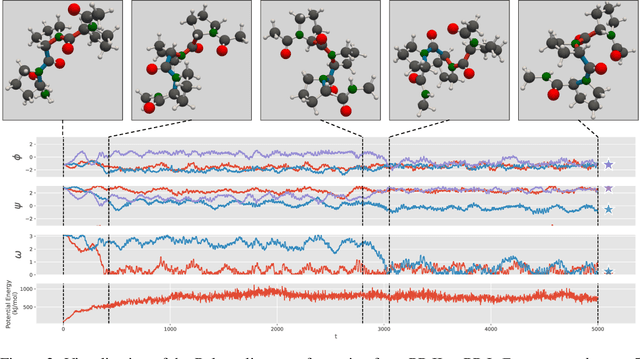
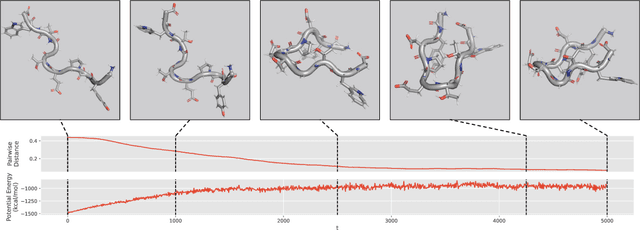
Abstract:We consider the problem of Sampling Transition Paths. Given two metastable conformational states of a molecular system, eg. a folded and unfolded protein, we aim to sample the most likely transition path between the two states. Sampling such a transition path is computationally expensive due to the existence of high free energy barriers between the two states. To circumvent this, previous work has focused on simplifying the trajectories to occur along specific molecular descriptors called Collective Variables (CVs). However, finding CVs is not trivial and requires chemical intuition. For larger molecules, where intuition is not sufficient, using these CV-based methods biases the transition along possibly irrelevant dimensions. Instead, this work proposes a method for sampling transition paths that consider the entire geometry of the molecules. To achieve this, we first relate the problem to recent work on the Schrodinger bridge problem and stochastic optimal control. Using this relation, we construct a method that takes into account important characteristics of molecular systems such as second-order dynamics and invariance to rotations and translations. We demonstrate our method on the commonly studied Alanine Dipeptide, but also consider larger proteins such as Polyproline and Chignolin.
Complex-Valued Autoencoders for Object Discovery
Apr 05, 2022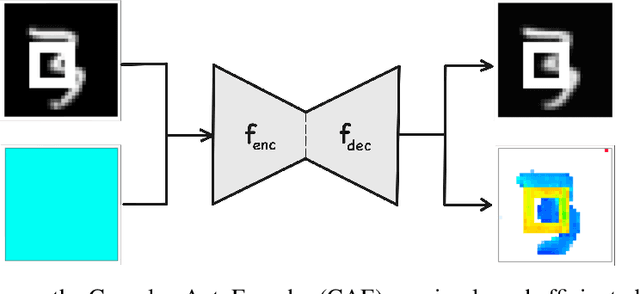
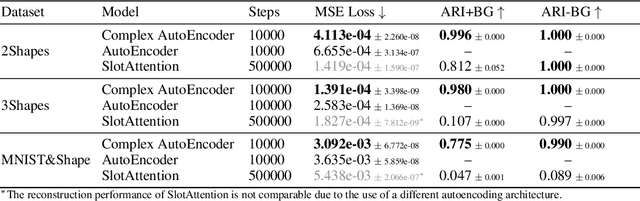
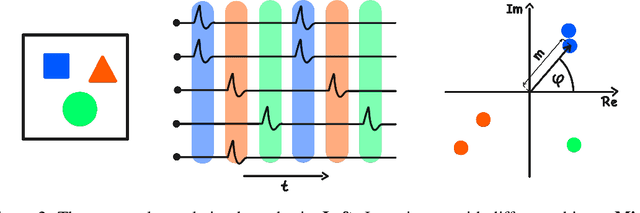
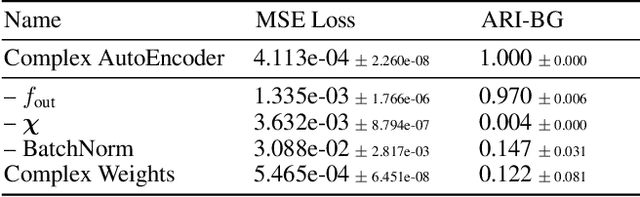
Abstract:Object-centric representations form the basis of human perception and enable us to reason about the world and to systematically generalize to new settings. Currently, most machine learning work on unsupervised object discovery focuses on slot-based approaches, which explicitly separate the latent representations of individual objects. While the result is easily interpretable, it usually requires the design of involved architectures. In contrast to this, we propose a distributed approach to object-centric representations: the Complex AutoEncoder. Following a coding scheme theorized to underlie object representations in biological neurons, its complex-valued activations represent two messages: their magnitudes express the presence of a feature, while the relative phase differences between neurons express which features should be bound together to create joint object representations. We show that this simple and efficient approach achieves better reconstruction performance than an equivalent real-valued autoencoder on simple multi-object datasets. Additionally, we show that it achieves competitive unsupervised object discovery performance to a SlotAttention model on two datasets, and manages to disentangle objects in a third dataset where SlotAttention fails - all while being 7-70 times faster to train.
Equivariant Diffusion for Molecule Generation in 3D
Mar 31, 2022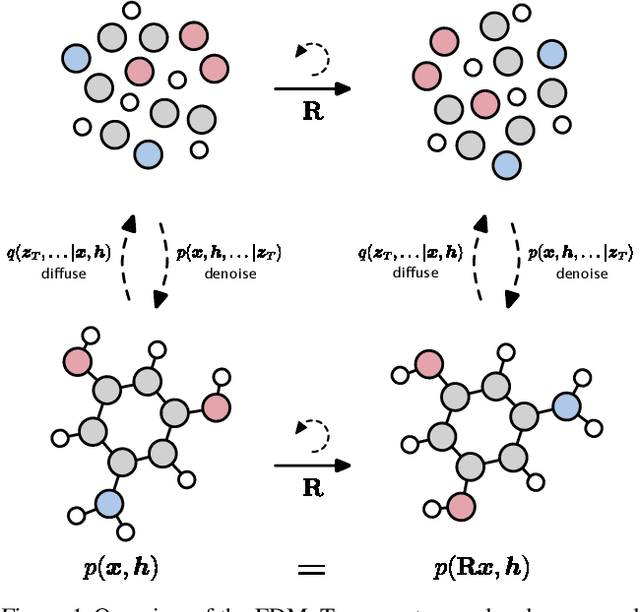



Abstract:This work introduces a diffusion model for molecule generation in 3D that is equivariant to Euclidean transformations. Our E(3) Equivariant Diffusion Model (EDM) learns to denoise a diffusion process with an equivariant network that jointly operates on both continuous (atom coordinates) and categorical features (atom types). In addition, we provide a probabilistic analysis which admits likelihood computation of molecules using our model. Experimentally, the proposed method significantly outperforms previous 3D molecular generative methods regarding the quality of generated samples and efficiency at training time.
Adversarial Defense via Image Denoising with Chaotic Encryption
Mar 19, 2022

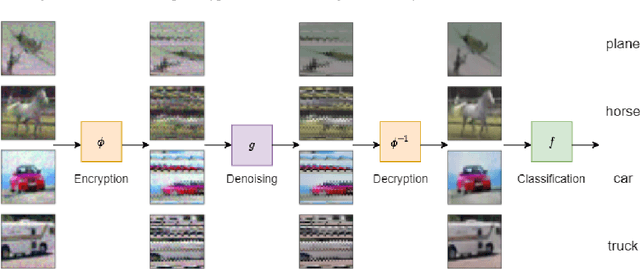
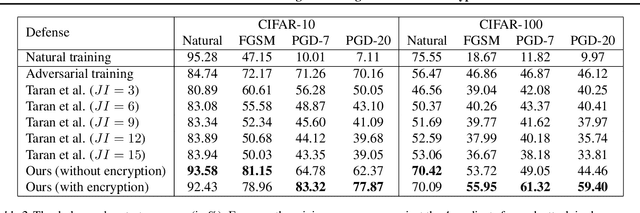
Abstract:In the literature on adversarial examples, white box and black box attacks have received the most attention. The adversary is assumed to have either full (white) or no (black) access to the defender's model. In this work, we focus on the equally practical gray box setting, assuming an attacker has partial information. We propose a novel defense that assumes everything but a private key will be made available to the attacker. Our framework uses an image denoising procedure coupled with encryption via a discretized Baker map. Extensive testing against adversarial images (e.g. FGSM, PGD) crafted using various gradients shows that our defense achieves significantly better results on CIFAR-10 and CIFAR-100 than the state-of-the-art gray box defenses in both natural and adversarial accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge