Max Tian
Using Scaling Laws for Data Source Utility Estimation in Domain-Specific Pre-Training
Jul 29, 2025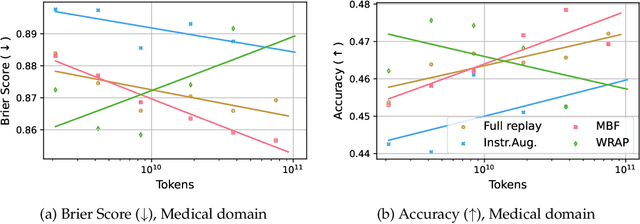
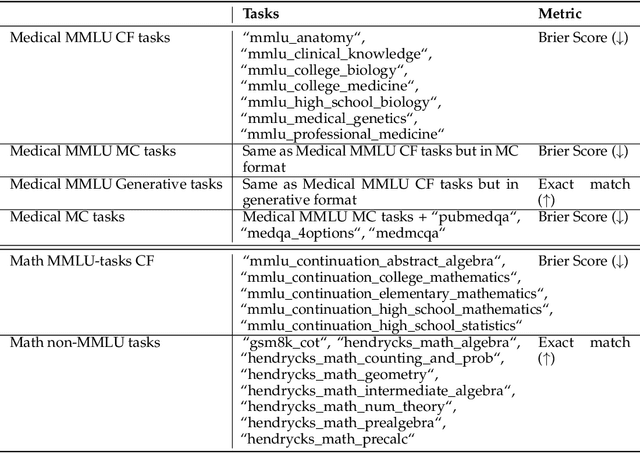
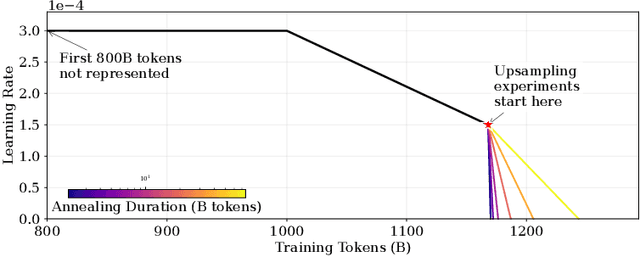
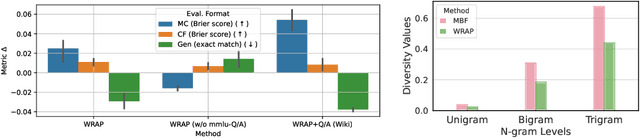
Abstract:We introduce a framework for optimizing domain-specific dataset construction in foundation model training. Specifically, we seek a cost-efficient way to estimate the quality of data sources (e.g. synthetically generated or filtered web data, etc.) in order to make optimal decisions about resource allocation for data sourcing from these sources for the stage two pre-training phase, aka annealing, with the goal of specializing a generalist pre-trained model to specific domains. Our approach extends the usual point estimate approaches, aka micro-annealing, to estimating scaling laws by performing multiple annealing runs of varying compute spent on data curation and training. This addresses a key limitation in prior work, where reliance on point estimates for data scaling decisions can be misleading due to the lack of rank invariance across compute scales -- a phenomenon we confirm in our experiments. By systematically analyzing performance gains relative to acquisition costs, we find that scaling curves can be estimated for different data sources. Such scaling laws can inform cost effective resource allocation across different data acquisition methods (e.g. synthetic data), data sources (e.g. user or web data) and available compute resources. We validate our approach through experiments on a pre-trained model with 7 billion parameters. We adapt it to: a domain well-represented in the pre-training data -- the medical domain, and a domain underrepresented in the pretraining corpora -- the math domain. We show that one can efficiently estimate the scaling behaviors of a data source by running multiple annealing runs, which can lead to different conclusions, had one used point estimates using the usual micro-annealing technique instead. This enables data-driven decision-making for selecting and optimizing data sources.
StarCoder 2 and The Stack v2: The Next Generation
Feb 29, 2024



Abstract:The BigCode project, an open-scientific collaboration focused on the responsible development of Large Language Models for Code (Code LLMs), introduces StarCoder2. In partnership with Software Heritage (SWH), we build The Stack v2 on top of the digital commons of their source code archive. Alongside the SWH repositories spanning 619 programming languages, we carefully select other high-quality data sources, such as GitHub pull requests, Kaggle notebooks, and code documentation. This results in a training set that is 4x larger than the first StarCoder dataset. We train StarCoder2 models with 3B, 7B, and 15B parameters on 3.3 to 4.3 trillion tokens and thoroughly evaluate them on a comprehensive set of Code LLM benchmarks. We find that our small model, StarCoder2-3B, outperforms other Code LLMs of similar size on most benchmarks, and also outperforms StarCoderBase-15B. Our large model, StarCoder2- 15B, significantly outperforms other models of comparable size. In addition, it matches or outperforms CodeLlama-34B, a model more than twice its size. Although DeepSeekCoder- 33B is the best-performing model at code completion for high-resource languages, we find that StarCoder2-15B outperforms it on math and code reasoning benchmarks, as well as several low-resource languages. We make the model weights available under an OpenRAIL license and ensure full transparency regarding the training data by releasing the SoftWare Heritage persistent IDentifiers (SWHIDs) of the source code data.
An Experimental Evaluation of Transformer-based Language Models in the Biomedical Domain
Dec 31, 2020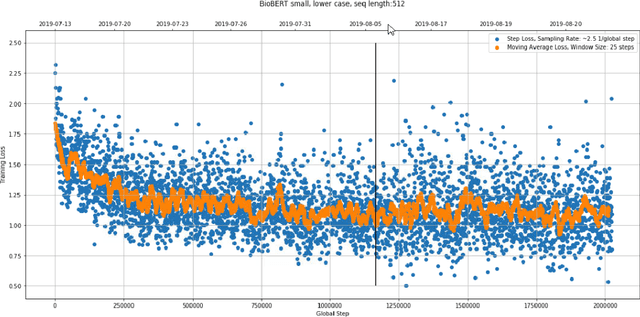
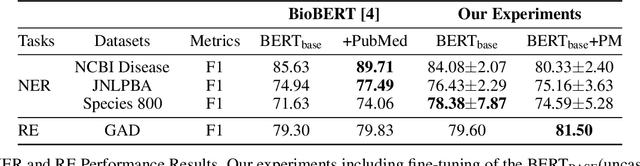
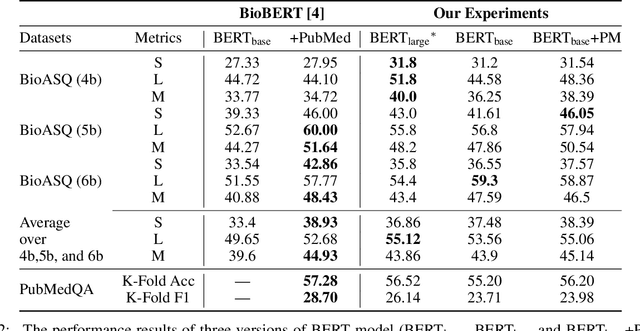
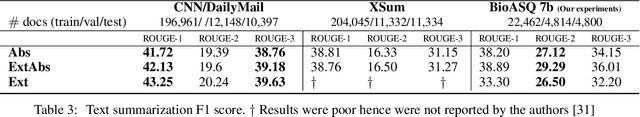
Abstract:With the growing amount of text in health data, there have been rapid advances in large pre-trained models that can be applied to a wide variety of biomedical tasks with minimal task-specific modifications. Emphasizing the cost of these models, which renders technical replication challenging, this paper summarizes experiments conducted in replicating BioBERT and further pre-training and careful fine-tuning in the biomedical domain. We also investigate the effectiveness of domain-specific and domain-agnostic pre-trained models across downstream biomedical NLP tasks. Our finding confirms that pre-trained models can be impactful in some downstream NLP tasks (QA and NER) in the biomedical domain; however, this improvement may not justify the high cost of domain-specific pre-training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge