Marleen de Bruijne
for the ALFA study
Crowdsourcing Airway Annotations in Chest Computed Tomography Images
Nov 20, 2020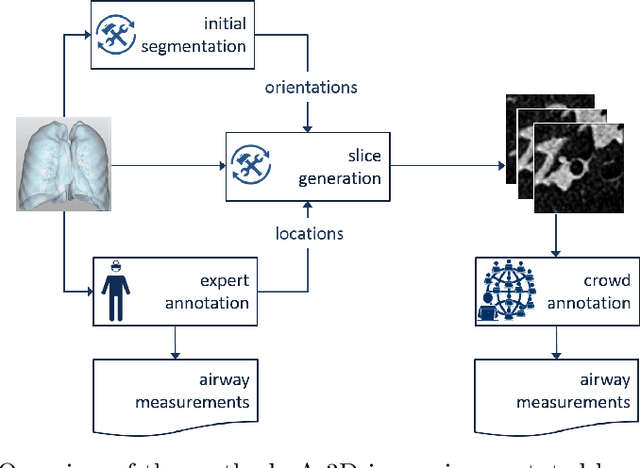
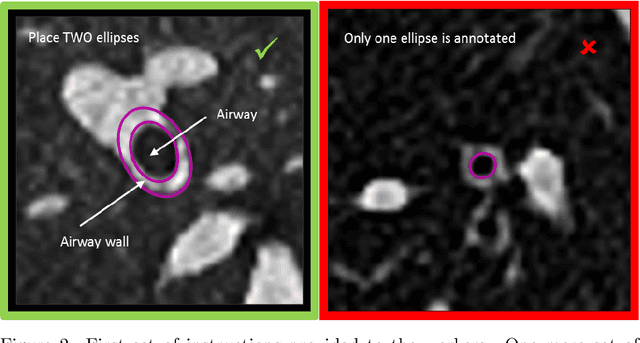
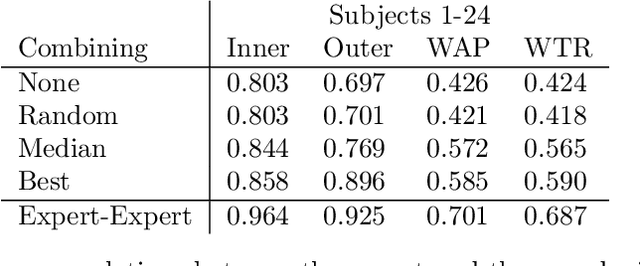
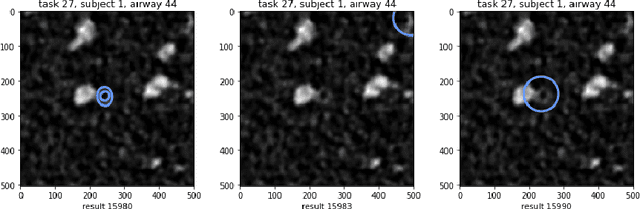
Abstract:Measuring airways in chest computed tomography (CT) scans is important for characterizing diseases such as cystic fibrosis, yet very time-consuming to perform manually. Machine learning algorithms offer an alternative, but need large sets of annotated scans for good performance. We investigate whether crowdsourcing can be used to gather airway annotations. We generate image slices at known locations of airways in 24 subjects and request the crowd workers to outline the airway lumen and airway wall. After combining multiple crowd workers, we compare the measurements to those made by the experts in the original scans. Similar to our preliminary study, a large portion of the annotations were excluded, possibly due to workers misunderstanding the instructions. After excluding such annotations, moderate to strong correlations with the expert can be observed, although these correlations are slightly lower than inter-expert correlations. Furthermore, the results across subjects in this study are quite variable. Although the crowd has potential in annotating airways, further development is needed for it to be robust enough for gathering annotations in practice. For reproducibility, data and code are available online: \url{http://github.com/adriapr/crowdairway.git}.
Medical Imaging with Deep Learning: MIDL 2020 -- Short Paper Track
Jun 29, 2020Abstract:This compendium gathers all the accepted extended abstracts from the Third International Conference on Medical Imaging with Deep Learning (MIDL 2020), held in Montreal, Canada, 6-9 July 2020. Note that only accepted extended abstracts are listed here, the Proceedings of the MIDL 2020 Full Paper Track are published in the Proceedings of Machine Learning Research (PMLR).
Region-of-interest guided Supervoxel Inpainting for Self-supervision
Jun 26, 2020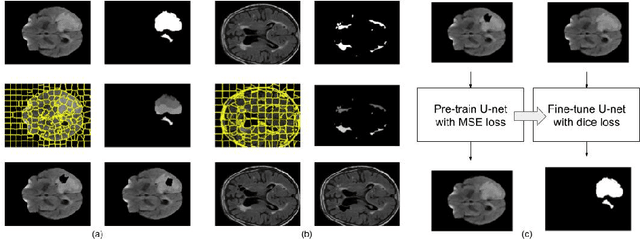
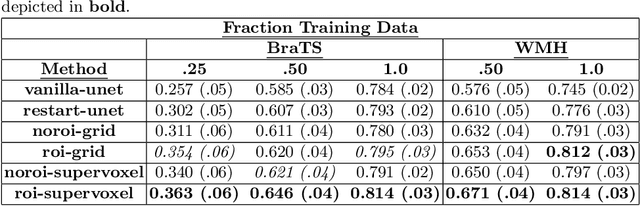
Abstract:Self-supervised learning has proven to be invaluable in making best use of all of the available data in biomedical image segmentation. One particularly simple and effective mechanism to achieve self-supervision is inpainting, the task of predicting arbitrary missing areas based on the rest of an image. In this work, we focus on image inpainting as the self-supervised proxy task, and propose two novel structural changes to further enhance the performance of a deep neural network. We guide the process of generating images to inpaint by using supervoxel-based masking instead of random masking, and also by focusing on the area to be segmented in the primary task, which we term as the region-of-interest. We postulate that these additions force the network to learn semantics that are more attuned to the primary task, and test our hypotheses on two applications: brain tumour and white matter hyperintensities segmentation. We empirically show that our proposed approach consistently outperforms both supervised CNNs, without any self-supervision, and conventional inpainting-based self-supervision methods on both large and small training set sizes.
DS6: Deformation-aware learning for small vessel segmentation with small, imperfectly labeled dataset
Jun 18, 2020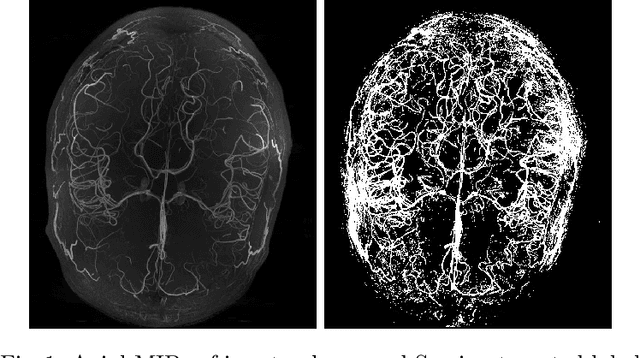
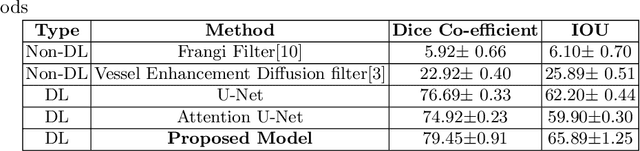
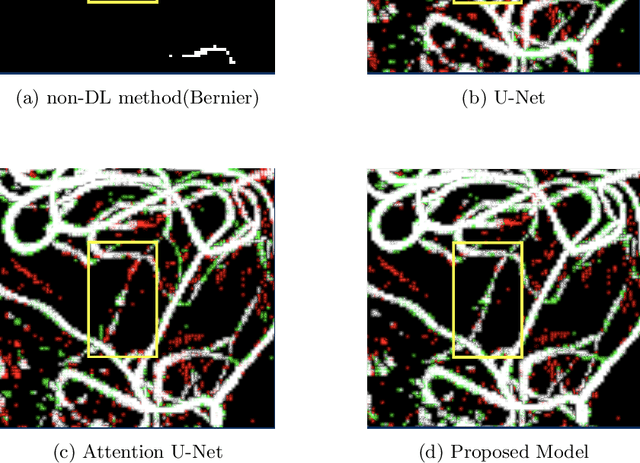

Abstract:Originating from the initial segment of the middle cerebral artery of the human brain, Lenticulostriate Arteries (LSA) are a collection of perforating vessels that supply blood to the basal ganglia region. With the advancement of 7 Tesla scanner, we are able to detect these LSA which are linked to Small Vessel Diseases(SVD) and potentially a cause for neurodegenerative diseases. Segmentation of LSA with traditional approaches like Frangi or semi-automated/manual techniques can depict medium to large vessels but fail to depict the small vessels. Also, semi-automated/manual approaches are time-consuming. In this paper, we put forth a study that incorporates deep learning techniques to automatically segment these LSA using 3D 7 Tesla Time-of-fight Magnetic Resonance Angiogram images. The algorithm is trained and evaluated on a small dataset of 11 volumes. Deep learning models based on Multi-Scale Supervision U-Net accompanied by elastic deformations to give equivariance to the model, were utilized for the vessel segmentation using semi-automated labeled images. We make a qualitative analysis of the output with the original image and also on imperfect semi-manual labels to confirm the presence and continuity of small vessels.
Adversarial Attack Vulnerability of Medical Image Analysis Systems: Unexplored Factors
Jun 12, 2020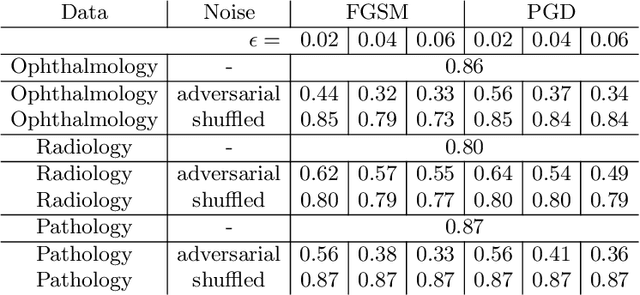
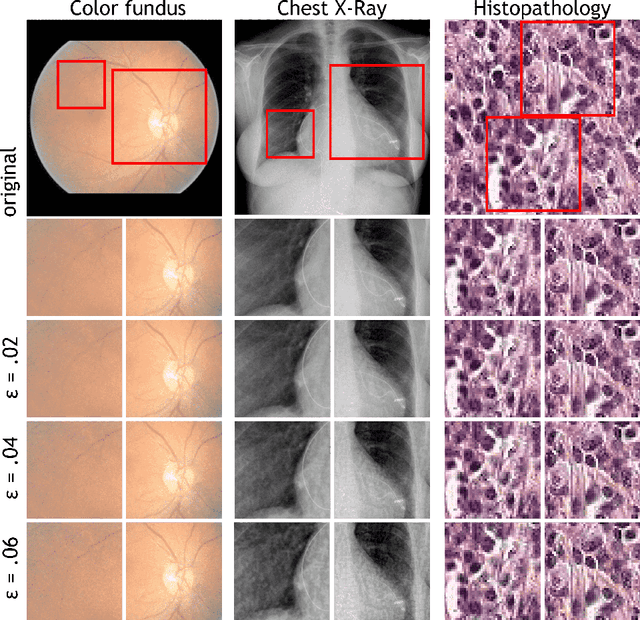
Abstract:Adversarial attacks are considered a potentially serious security threat for machine learning systems. Medical image analysis (MedIA) systems have recently been argued to be particularly vulnerable to adversarial attacks due to strong financial incentives. In this paper, we study several previously unexplored factors affecting adversarial attack vulnerability of deep learning MedIA systems in three medical domains: ophthalmology, radiology and pathology. Firstly, we study the effect of varying the degree of adversarial perturbation on the attack performance and its visual perceptibility. Secondly, we study how pre-training on a public dataset (ImageNet) affects the models' vulnerability to attacks. Thirdly, we study the influence of data and model architecture disparity between target and attacker models. Our experiments show that the degree of perturbation significantly affects both performance and human perceptibility of attacks. Pre-training may dramatically increase the transfer of adversarial examples; the larger the performance gain achieved by pre-training, the larger the transfer. Finally, disparity in data and/or model architecture between target and attacker models substantially decreases the success of attacks. We believe that these factors should be considered when designing cybersecurity-critical MedIA systems, as well as kept in mind when evaluating their vulnerability to adversarial attacks.
Spectral Data Augmentation Techniques to quantify Lung Pathology from CT-images
Apr 24, 2020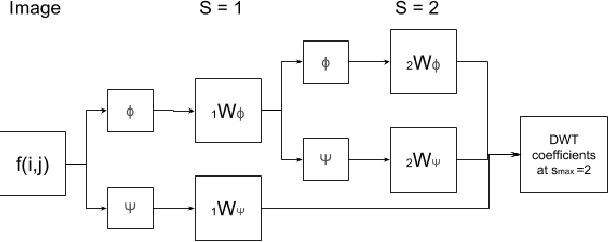
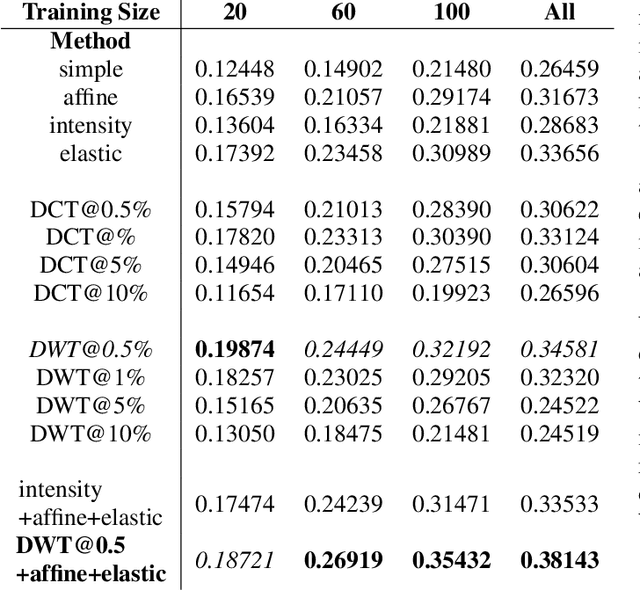
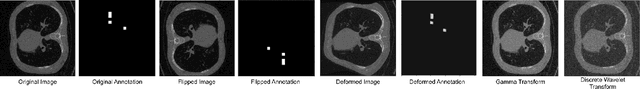
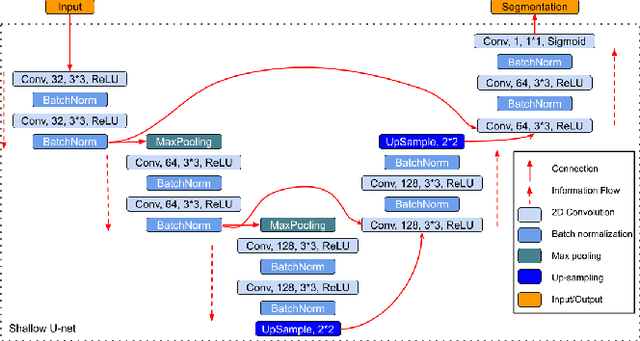
Abstract:Data augmentation is of paramount importance in biomedical image processing tasks, characterized by inadequate amounts of labelled data, to best use all of the data that is present. In-use techniques range from intensity transformations and elastic deformations, to linearly combining existing data points to make new ones. In this work, we propose the use of spectral techniques for data augmentation, using the discrete cosine and wavelet transforms. We empirically evaluate our approaches on a CT texture analysis task to detect abnormal lung-tissue in patients with cystic fibrosis. Empirical experiments show that the proposed spectral methods perform favourably as compared to the existing methods. When used in combination with existing methods, our proposed approach can increase the relative minor class segmentation performance by 44.1% over a simple replication baseline.
When Weak Becomes Strong: Robust Quantification of White Matter Hyperintensities in Brain MRI scans
Apr 12, 2020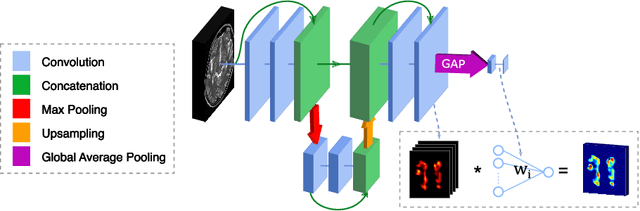
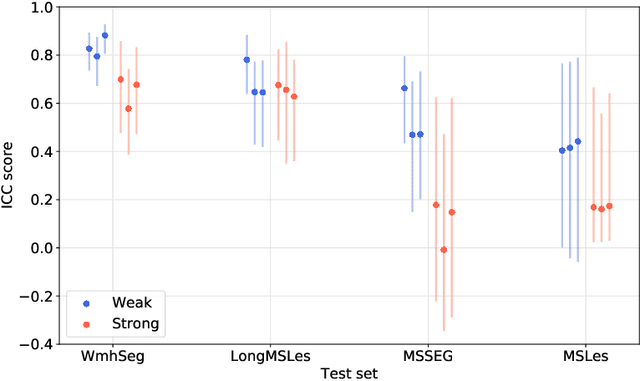
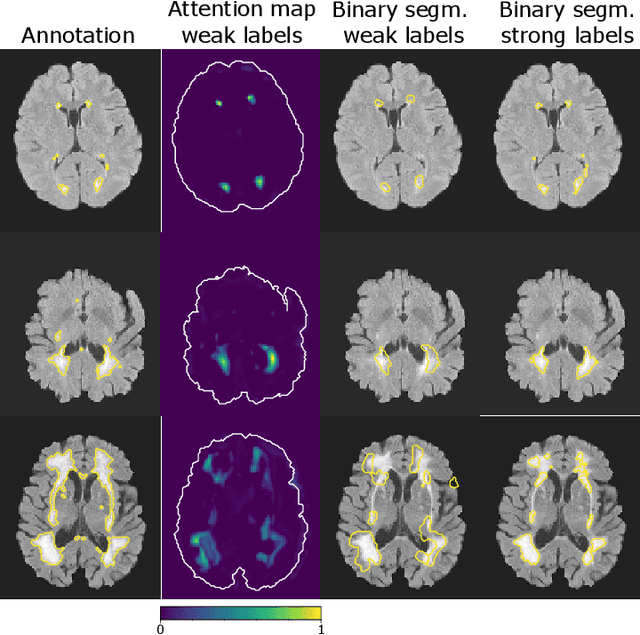
Abstract:To measure the volume of specific image structures, a typical approach is to first segment those structures using a neural network trained on voxel-wise (strong) labels and subsequently compute the volume from the segmentation. A more straightforward approach would be to predict the volume directly using a neural network based regression approach, trained on image-level (weak) labels indicating volume. In this article, we compared networks optimized with weak and strong labels, and study their ability to generalize to other datasets. We experimented with white matter hyperintensity (WMH) volume prediction in brain MRI scans. Neural networks were trained on a large local dataset and their performance was evaluated on four independent public datasets. We showed that networks optimized using only weak labels reflecting WMH volume generalized better for WMH volume prediction than networks optimized with voxel-wise segmentations of WMH. The attention maps of networks trained with weak labels did not seem to delineate WMHs, but highlighted instead areas with smooth contours around or near WMHs. By correcting for possible confounders we showed that networks trained on weak labels may have learnt other meaningful features that are more suited to generalization to unseen data. Our results suggest that for imaging biomarkers that can be derived from segmentations, training networks to predict the biomarker directly may provide more robust results than solving an intermediate segmentation step.
Automated Estimation of the Spinal Curvature via Spine Centerline Extraction with Ensembles of Cascaded Neural Networks
Dec 11, 2019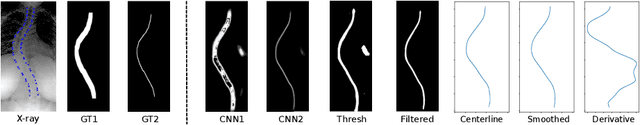

Abstract:Scoliosis is a condition defined by an abnormal spinal curvature. For diagnosis and treatment planning of scoliosis, spinal curvature can be estimated using Cobb angles. We propose an automated method for the estimation of Cobb angles from X-ray scans. First, the centerline of the spine was segmented using a cascade of two convolutional neural networks. After smoothing the centerline, Cobb angles were automatically estimated using the derivative of the centerline. We evaluated the results using the mean absolute error and the average symmetric mean absolute percentage error between the manual assessment by experts and the automated predictions. For optimization, we used 609 X-ray scans from the London Health Sciences Center, and for evaluation, we participated in the international challenge "Accurate Automated Spinal Curvature Estimation, MICCAI 2019" (100 scans). On the challenge's test set, we obtained an average symmetric mean absolute percentage error of 22.96.
Semi-Supervised Medical Image Segmentation via Learning Consistency under Transformations
Nov 04, 2019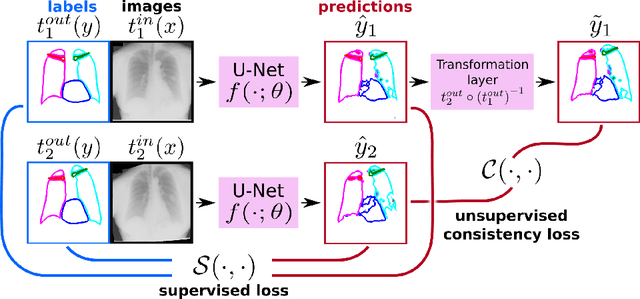
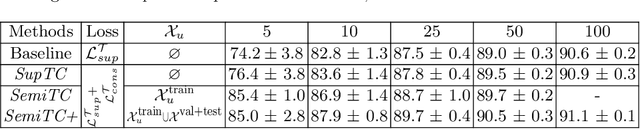
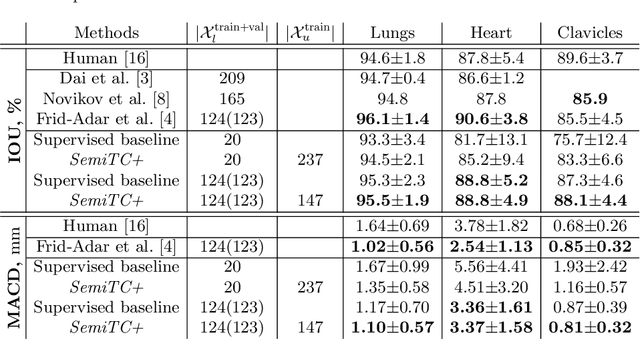
Abstract:The scarcity of labeled data often limits the application of supervised deep learning techniques for medical image segmentation. This has motivated the development of semi-supervised techniques that learn from a mixture of labeled and unlabeled images. In this paper, we propose a novel semi-supervised method that, in addition to supervised learning on labeled training images, learns to predict segmentations consistent under a given class of transformations on both labeled and unlabeled images. More specifically, in this work we explore learning equivariance to elastic deformations. We implement this through: 1) a Siamese architecture with two identical branches, each of which receives a differently transformed image, and 2) a composite loss function with a supervised segmentation loss term and an unsupervised term that encourages segmentation consistency between the predictions of the two branches. We evaluate the method on a public dataset of chest radiographs with segmentations of anatomical structures using 5-fold cross-validation. The proposed method reaches significantly higher segmentation accuracy compared to supervised learning. This is due to learning transformation consistency on both labeled and unlabeled images, with the latter contributing the most. We achieve the performance comparable to state-of-the-art chest X-ray segmentation methods while using substantially fewer labeled images.
APIR-Net: Autocalibrated Parallel Imaging Reconstruction using a Neural Network
Sep 19, 2019



Abstract:Deep learning has been successfully demonstrated in MRI reconstruction of accelerated acquisitions. However, its dependence on representative training data limits the application across different contrasts, anatomies, or image sizes. To address this limitation, we propose an unsupervised, auto-calibrated k-space completion method, based on a uniquely designed neural network that reconstructs the full k-space from an undersampled k-space, exploiting the redundancy among the multiple channels in the receive coil in a parallel imaging acquisition. To achieve this, contrary to common convolutional network approaches, the proposed network has a decreasing number of feature maps of constant size. In contrast to conventional parallel imaging methods such as GRAPPA that estimate the prediction kernel from the fully sampled autocalibration signals in a linear way, our method is able to learn nonlinear relations between sampled and unsampled positions in k-space. The proposed method was compared to the start-of-the-art ESPIRiT and RAKI methods in terms of noise amplification and visual image quality in both phantom and in-vivo experiments. The experiments indicate that APIR-Net provides a promising alternative to the conventional parallel imaging methods, and results in improved image quality especially for low SNR acquisitions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge