Oliver Speck
SMILE-UHURA Challenge -- Small Vessel Segmentation at Mesoscopic Scale from Ultra-High Resolution 7T Magnetic Resonance Angiograms
Nov 14, 2024Abstract:The human brain receives nutrients and oxygen through an intricate network of blood vessels. Pathology affecting small vessels, at the mesoscopic scale, represents a critical vulnerability within the cerebral blood supply and can lead to severe conditions, such as Cerebral Small Vessel Diseases. The advent of 7 Tesla MRI systems has enabled the acquisition of higher spatial resolution images, making it possible to visualise such vessels in the brain. However, the lack of publicly available annotated datasets has impeded the development of robust, machine learning-driven segmentation algorithms. To address this, the SMILE-UHURA challenge was organised. This challenge, held in conjunction with the ISBI 2023, in Cartagena de Indias, Colombia, aimed to provide a platform for researchers working on related topics. The SMILE-UHURA challenge addresses the gap in publicly available annotated datasets by providing an annotated dataset of Time-of-Flight angiography acquired with 7T MRI. This dataset was created through a combination of automated pre-segmentation and extensive manual refinement. In this manuscript, sixteen submitted methods and two baseline methods are compared both quantitatively and qualitatively on two different datasets: held-out test MRAs from the same dataset as the training data (with labels kept secret) and a separate 7T ToF MRA dataset where both input volumes and labels are kept secret. The results demonstrate that most of the submitted deep learning methods, trained on the provided training dataset, achieved reliable segmentation performance. Dice scores reached up to 0.838 $\pm$ 0.066 and 0.716 $\pm$ 0.125 on the respective datasets, with an average performance of up to 0.804 $\pm$ 0.15.
SPOCKMIP: Segmentation of Vessels in MRAs with Enhanced Continuity using Maximum Intensity Projection as Loss
Jul 11, 2024



Abstract:Identification of vessel structures of different sizes in biomedical images is crucial in the diagnosis of many neurodegenerative diseases. However, the sparsity of good-quality annotations of such images makes the task of vessel segmentation challenging. Deep learning offers an efficient way to segment vessels of different sizes by learning their high-level feature representations and the spatial continuity of such features across dimensions. Semi-supervised patch-based approaches have been effective in identifying small vessels of one to two voxels in diameter. This study focuses on improving the segmentation quality by considering the spatial correlation of the features using the Maximum Intensity Projection~(MIP) as an additional loss criterion. Two methods are proposed with the incorporation of MIPs of label segmentation on the single~(z-axis) and multiple perceivable axes of the 3D volume. The proposed MIP-based methods produce segmentations with improved vessel continuity, which is evident in visual examinations of ROIs. Patch-based training is improved by introducing an additional loss term, MIP loss, to penalise the predicted discontinuity of vessels. A training set of 14 volumes is selected from the StudyForrest dataset comprising of 18 7-Tesla 3D Time-of-Flight~(ToF) Magnetic Resonance Angiography (MRA) images. The generalisation performance of the method is evaluated using the other unseen volumes in the dataset. It is observed that the proposed method with multi-axes MIP loss produces better quality segmentations with a median Dice of $80.245 \pm 0.129$. Also, the method with single-axis MIP loss produces segmentations with a median Dice of $79.749 \pm 0.109$. Furthermore, a visual comparison of the ROIs in the predicted segmentation reveals a significant improvement in the continuity of the vessels when MIP loss is incorporated into training.
PULASki: Learning inter-rater variability using statistical distances to improve probabilistic segmentation
Dec 25, 2023



Abstract:In the domain of medical imaging, many supervised learning based methods for segmentation face several challenges such as high variability in annotations from multiple experts, paucity of labelled data and class imbalanced datasets. These issues may result in segmentations that lack the requisite precision for clinical analysis and can be misleadingly overconfident without associated uncertainty quantification. We propose the PULASki for biomedical image segmentation that accurately captures variability in expert annotations, even in small datasets. Our approach makes use of an improved loss function based on statistical distances in a conditional variational autoencoder structure (Probabilistic UNet), which improves learning of the conditional decoder compared to the standard cross-entropy particularly in class imbalanced problems. We analyse our method for two structurally different segmentation tasks (intracranial vessel and multiple sclerosis (MS) lesion) and compare our results to four well-established baselines in terms of quantitative metrics and qualitative output. Empirical results demonstrate the PULASKi method outperforms all baselines at the 5\% significance level. The generated segmentations are shown to be much more anatomically plausible than in the 2D case, particularly for the vessel task. Our method can also be applied to a wide range of multi-label segmentation tasks and and is useful for downstream tasks such as hemodynamic modelling (computational fluid dynamics and data assimilation), clinical decision making, and treatment planning.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Complex Network for Complex Problems: A comparative study of CNN and Complex-valued CNN
Feb 09, 2023



Abstract:Neural networks, especially convolutional neural networks (CNN), are one of the most common tools these days used in computer vision. Most of these networks work with real-valued data using real-valued features. Complex-valued convolutional neural networks (CV-CNN) can preserve the algebraic structure of complex-valued input data and have the potential to learn more complex relationships between the input and the ground-truth. Although some comparisons of CNNs and CV-CNNs for different tasks have been performed in the past, a large-scale investigation comparing different models operating on different tasks has not been conducted. Furthermore, because complex features contain both real and imaginary components, CV-CNNs have double the number of trainable parameters as real-valued CNNs in terms of the actual number of trainable parameters. Whether or not the improvements in performance with CV-CNN observed in the past have been because of the complex features or just because of having double the number of trainable parameters has not yet been explored. This paper presents a comparative study of CNN, CNNx2 (CNN with double the number of trainable parameters as the CNN), and CV-CNN. The experiments were performed using seven models for two different tasks - brain tumour classification and segmentation in brain MRIs. The results have revealed that the CV-CNN models outperformed the CNN and CNNx2 models.
Liver Segmentation in Time-resolved C-arm CT Volumes Reconstructed from Dynamic Perfusion Scans using Time Separation Technique
Feb 09, 2023



Abstract:Perfusion imaging is a valuable tool for diagnosing and treatment planning for liver tumours. The time separation technique (TST) has been successfully used for modelling C-arm cone-beam computed tomography (CBCT) perfusion data. The reconstruction can be accompanied by the segmentation of the liver - for better visualisation and for generating comprehensive perfusion maps. Recently introduced Turbolift learning has been seen to perform well while working with TST reconstructions, but has not been explored for the time-resolved volumes (TRV) estimated out of TST reconstructions. The segmentation of the TRVs can be useful for tracking the movement of the liver over time. This research explores this possibility by training the multi-scale attention UNet of Turbolift learning at its third stage on the TRVs and shows the robustness of Turbolift learning since it can even work efficiently with the TRVs, resulting in a Dice score of 0.864$\pm$0.004.
Liver Segmentation using Turbolift Learning for CT and Cone-beam C-arm Perfusion Imaging
Jul 20, 2022
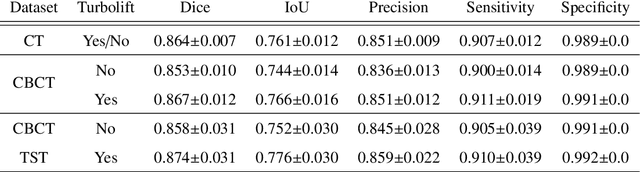
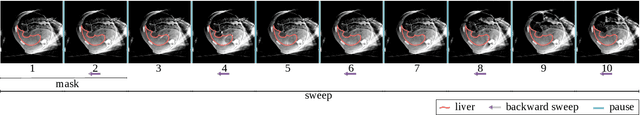
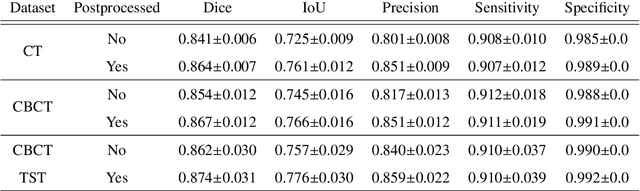
Abstract:Model-based reconstruction employing the time separation technique (TST) was found to improve dynamic perfusion imaging of the liver using C-arm cone-beam computed tomography (CBCT). To apply TST using prior knowledge extracted from CT perfusion data, the liver should be accurately segmented from the CT scans. Reconstructions of primary and model-based CBCT data need to be segmented for proper visualisation and interpretation of perfusion maps. This research proposes Turbolift learning, which trains a modified version of the multi-scale Attention UNet on different liver segmentation tasks serially, following the order of the trainings CT, CBCT, CBCT TST - making the previous trainings act as pre-training stages for the subsequent ones - addressing the problem of limited number of datasets for training. For the final task of liver segmentation from CBCT TST, the proposed method achieved an overall Dice scores of 0.874$\pm$0.031 and 0.905$\pm$0.007 in 6-fold and 4-fold cross-validation experiments, respectively - securing statistically significant improvements over the model, which was trained only for that task. Experiments revealed that Turbolift not only improves the overall performance of the model but also makes it robust against artefacts originating from the embolisation materials and truncation artefacts. Additionally, in-depth analyses confirmed the order of the segmentation tasks. This paper shows the potential of segmenting the liver from CT, CBCT, and CBCT TST, learning from the available limited training data, which can possibly be used in the future for the visualisation and evaluation of the perfusion maps for the treatment evaluation of liver diseases.
Automated SSIM Regression for Detection and Quantification of Motion Artefacts in Brain MR Images
Jun 14, 2022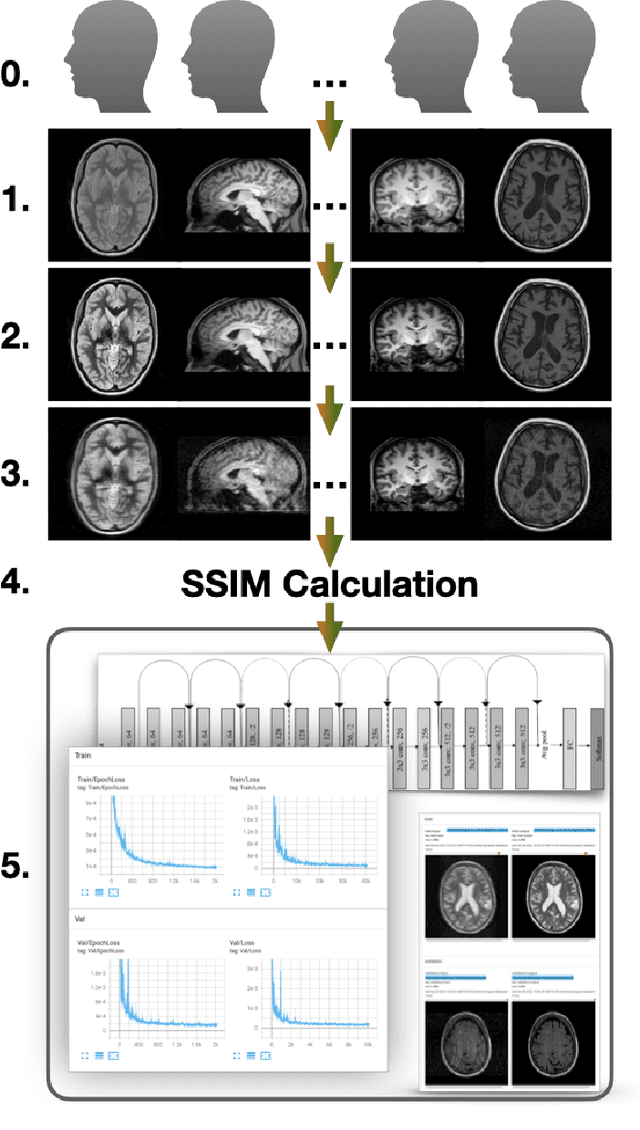
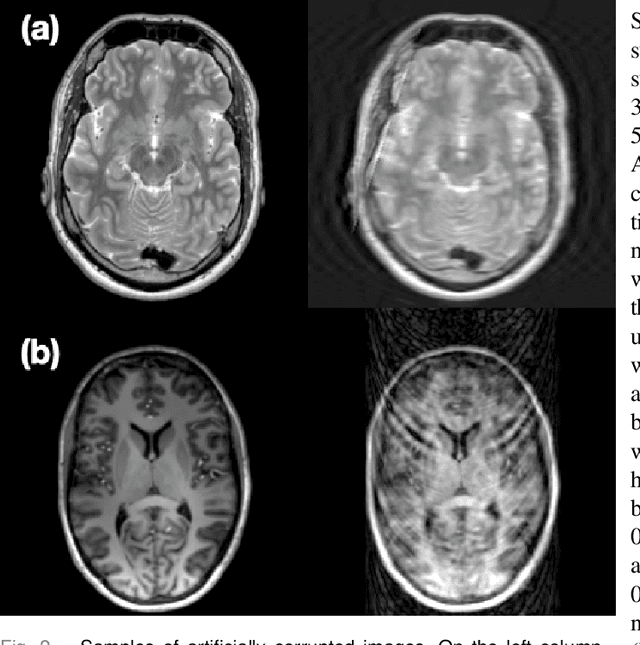
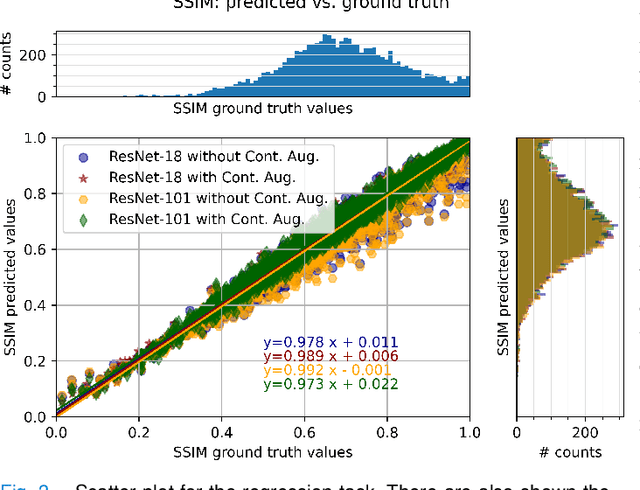
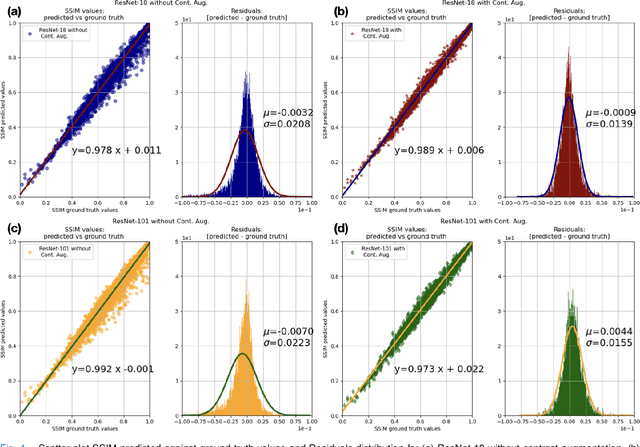
Abstract:Motion artefacts in magnetic resonance brain images are a crucial issue. The assessment of MR image quality is fundamental before proceeding with the clinical diagnosis. If the motion artefacts alter a correct delineation of structure and substructures of the brain, lesions, tumours and so on, the patients need to be re-scanned. Otherwise, neuro-radiologists could report an inaccurate or incorrect diagnosis. The first step right after scanning a patient is the "\textit{image quality assessment}" in order to decide if the acquired images are diagnostically acceptable. An automated image quality assessment based on the structural similarity index (SSIM) regression through a residual neural network has been proposed here, with the possibility to perform also the classification in different groups - by subdividing with SSIM ranges. This method predicts SSIM values of an input image in the absence of a reference ground truth image. The networks were able to detect motion artefacts, and the best performance for the regression and classification task has always been achieved with ResNet-18 with contrast augmentation. Mean and standard deviation of residuals' distribution were $\mu=-0.0009$ and $\sigma=0.0139$, respectively. Whilst for the classification task in 3, 5 and 10 classes, the best accuracies were 97, 95 and 89\%, respectively. The obtained results show that the proposed method could be a tool in supporting neuro-radiologists and radiographers in evaluating the image quality before the diagnosis.
Weakly-supervised segmentation using inherently-explainable classification models and their application to brain tumour classification
Jun 10, 2022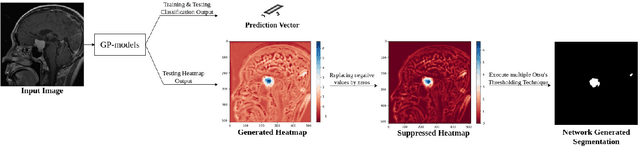
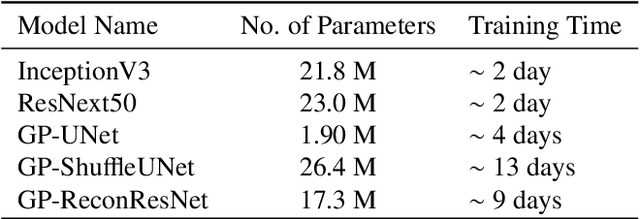
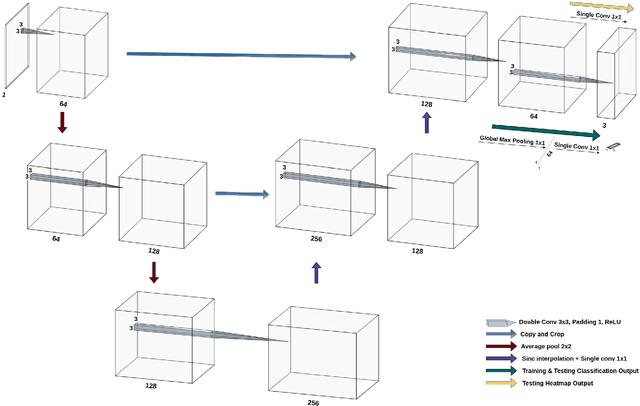
Abstract:Deep learning models have shown their potential for several applications. However, most of the models are opaque and difficult to trust due to their complex reasoning - commonly known as the black-box problem. Some fields, such as medicine, require a high degree of transparency to accept and adopt such technologies. Consequently, creating explainable/interpretable models or applying post-hoc methods on classifiers to build trust in deep learning models are required. Moreover, deep learning methods can be used for segmentation tasks, which typically require hard-to-obtain, time-consuming manually-annotated segmentation labels for training. This paper introduces three inherently-explainable classifiers to tackle both of these problems as one. The localisation heatmaps provided by the networks -- representing the models' focus areas and being used in classification decision-making -- can be directly interpreted, without requiring any post-hoc methods to derive information for model explanation. The models are trained by using the input image and only the classification labels as ground-truth in a supervised fashion - without using any information about the location of the region of interest (i.e. the segmentation labels), making the segmentation training of the models weakly-supervised through classification labels. The final segmentation is obtained by thresholding these heatmaps. The models were employed for the task of multi-class brain tumour classification using two different datasets, resulting in the best F1-score of 0.93 for the supervised classification task while securing a median Dice score of 0.67$\pm$0.08 for the weakly-supervised segmentation task. Furthermore, the obtained accuracy on a subset of tumour-only images outperformed the state-of-the-art glioma tumour grading binary classifiers with the best model achieving 98.7\% accuracy.
MICDIR: Multi-scale Inverse-consistent Deformable Image Registration using UNetMSS with Self-Constructing Graph Latent
Mar 08, 2022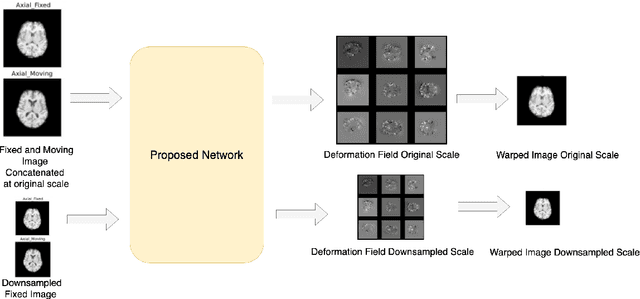
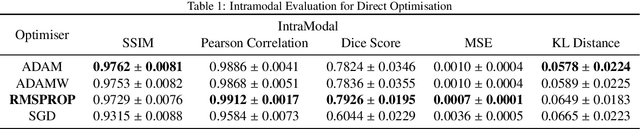
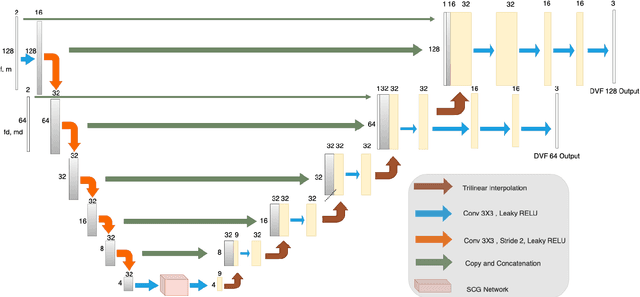
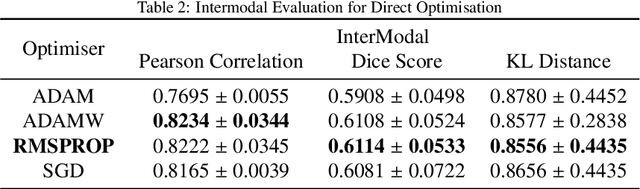
Abstract:Image registration is the process of bringing different images into a common coordinate system - a technique widely used in various applications of computer vision, such as remote sensing, image retrieval, and most commonly in medical imaging. Deep Learning based techniques have been applied successfully to tackle various complex medical image processing problems, including medical image registration. Over the years, several image registration techniques have been proposed using deep learning. Deformable image registration techniques such as Voxelmorph have been successful in capturing finer changes and providing smoother deformations. However, Voxelmorph, as well as ICNet and FIRE, do not explicitly encode global dependencies (i.e. the overall anatomical view of the supplied image) and therefore can not track large deformations. In order to tackle the aforementioned problems, this paper extends the Voxelmorph approach in three different ways. To improve the performance in case of small as well as large deformations, supervision of the model at different resolutions have been integrated using a multi-scale UNet. To support the network to learn and encode the minute structural co-relations of the given image-pairs, a self-constructing graph network (SCGNet) has been used as the latent of the multi-scale UNet - which can improve the learning process of the model and help the model to generalise better. And finally, to make the deformations inverse-consistent, cycle consistency loss has been employed. On the task of registration of brain MRIs, the proposed method achieved significant improvements over ANTs and VoxelMorph, obtaining a Dice score of 0.8013$\pm$0.0243 for intramodal and 0.6211$\pm$0.0309 for intermodal, while VoxelMorph achieved 0.7747$\pm$0.0260 and 0.6071$\pm$0.0510, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge