Marcos Matabuena
Distributional Random Forests for Complex Survey Designs on Reproducing Kernel Hilbert Spaces
Dec 09, 2025Abstract:We study estimation of the conditional law $P(Y|X=\mathbf{x})$ and continuous functionals $Ψ(P(Y|X=\mathbf{x}))$ when $Y$ takes values in a locally compact Polish space, $X \in \mathbb{R}^p$, and the observations arise from a complex survey design. We propose a survey-calibrated distributional random forest (SDRF) that incorporates complex-design features via a pseudo-population bootstrap, PSU-level honesty, and a Maximum Mean Discrepancy (MMD) split criterion computed from kernel mean embeddings of Hájek-type (design-weighted) node distributions. We provide a framework for analyzing forest-style estimators under survey designs; establish design consistency for the finite-population target and model consistency for the super-population target under explicit conditions on the design, kernel, resampling multipliers, and tree partitions. As far as we are aware, these are the first results on model-free estimation of conditional distributions under survey designs. Simulations under a stratified two-stage cluster design provide finite sample performance and demonstrate the statistical error price of ignoring the survey design. The broad applicability of SDRF is demonstrated using NHANES: We estimate the tolerance regions of the conditional joint distribution of two diabetes biomarkers, illustrating how distributional heterogeneity can support subgroup-specific risk profiling for diabetes mellitus in the U.S. population.
ROC Analysis with Covariate Adjustment Using Neural Network Models: Evaluating the Role of Age in the Physical Activity-Mortality Association
Oct 16, 2025Abstract:The receiver operating characteristic (ROC) curve and its summary measure, the Area Under the Curve (AUC), are well-established tools for evaluating the efficacy of biomarkers in biomedical studies. Compared to the traditional ROC curve, the covariate-adjusted ROC curve allows for individual evaluation of the biomarker. However, the use of machine learning models has rarely been explored in this context, despite their potential to develop more powerful and sophisticated approaches for biomarker evaluation. The goal of this paper is to propose a framework for neural network-based covariate-adjusted ROC modeling that allows flexible and nonlinear evaluation of the effectiveness of a biomarker to discriminate between two reference populations. The finite-sample performance of our method is investigated through extensive simulation tests under varying dependency structures between biomarkers, covariates, and referenced populations. The methodology is further illustrated in a clinically case study that assesses daily physical activity - measured as total activity time (TAC), a proxy for daily step count-as a biomarker to predict mortality at three, five and eight years. Analyzes stratified by sex and adjusted for age and BMI reveal distinct covariate effects on mortality outcomes. These results underscore the importance of covariate-adjusted modeling in biomarker evaluation and highlight TAC's potential as a functional capacity biomarker based on specific individual characteristics.
Model-Free Kernel Conformal Depth Measures Algorithm for Uncertainty Quantification in Regression Models in Separable Hilbert Spaces
Jun 10, 2025Abstract:Depth measures are powerful tools for defining level sets in emerging, non--standard, and complex random objects such as high-dimensional multivariate data, functional data, and random graphs. Despite their favorable theoretical properties, the integration of depth measures into regression modeling to provide prediction regions remains a largely underexplored area of research. To address this gap, we propose a novel, model-free uncertainty quantification algorithm based on conditional depth measures--specifically, conditional kernel mean embeddings and an integrated depth measure. These new algorithms can be used to define prediction and tolerance regions when predictors and responses are defined in separable Hilbert spaces. The use of kernel mean embeddings ensures faster convergence rates in prediction region estimation. To enhance the practical utility of the algorithms with finite samples, we also introduce a conformal prediction variant that provides marginal, non-asymptotic guarantees for the derived prediction regions. Additionally, we establish both conditional and unconditional consistency results, as well as fast convergence rates in certain homoscedastic settings. We evaluate the finite--sample performance of our model in extensive simulation studies involving various types of functional data and traditional Euclidean scenarios. Finally, we demonstrate the practical relevance of our approach through a digital health application related to physical activity, aiming to provide personalized recommendations
Continuous Temporal Learning of Probability Distributions via Neural ODEs with Applications in Continuous Glucose Monitoring Data
May 13, 2025Abstract:Modeling the continuous--time dynamics of probability distributions from time--dependent data samples is a fundamental problem in many fields, including digital health. The aim is to analyze how the distribution of a biomarker, such as glucose, evolves over time and how these changes may reflect the progression of chronic diseases such as diabetes. In this paper, we propose a novel probabilistic model based on a mixture of Gaussian distributions to capture how samples from a continuous-time stochastic process evolve over the time. To model potential distribution shifts over time, we introduce a time-dependent function parameterized by a Neural Ordinary Differential Equation (Neural ODE) and estimate it non--parametrically using the Maximum Mean Discrepancy (MMD). The proposed model is highly interpretable, detects subtle temporal shifts, and remains computationally efficient. Through simulation studies, we show that it performs competitively in terms of estimation accuracy against state-of-the-art, less interpretable methods such as normalized gradient--flows and non--parameteric kernel density estimators. Finally, we demonstrate the utility of our method on digital clinical--trial data, showing how the interventions alters the time-dependent distribution of glucose levels and enabling a rigorous comparison of control and treatment groups from novel mathematical and clinical perspectives.
Variable Selection Methods for Multivariate, Functional, and Complex Biomedical Data in the AI Age
Jan 12, 2025



Abstract:Many problems within personalized medicine and digital health rely on the analysis of continuous-time functional biomarkers and other complex data structures emerging from high-resolution patient monitoring. In this context, this work proposes new optimization-based variable selection methods for multivariate, functional, and even more general outcomes in metrics spaces based on best-subset selection. Our framework applies to several types of regression models, including linear, quantile, or non parametric additive models, and to a broad range of random responses, such as univariate, multivariate Euclidean data, functional, and even random graphs. Our analysis demonstrates that our proposed methodology outperforms state-of-the-art methods in accuracy and, especially, in speed-achieving several orders of magnitude improvement over competitors across various type of statistical responses as the case of mathematical functions. While our framework is general and is not designed for a specific regression and scientific problem, the article is self-contained and focuses on biomedical applications. In the clinical areas, serves as a valuable resource for professionals in biostatistics, statistics, and artificial intelligence interested in variable selection problem in this new technological AI-era.
Conformal Prediction in Dynamic Biological Systems
Sep 04, 2024



Abstract:Uncertainty quantification (UQ) is the process of systematically determining and characterizing the degree of confidence in computational model predictions. In the context of systems biology, especially with dynamic models, UQ is crucial because it addresses the challenges posed by nonlinearity and parameter sensitivity, allowing us to properly understand and extrapolate the behavior of complex biological systems. Here, we focus on dynamic models represented by deterministic nonlinear ordinary differential equations. Many current UQ approaches in this field rely on Bayesian statistical methods. While powerful, these methods often require strong prior specifications and make parametric assumptions that may not always hold in biological systems. Additionally, these methods face challenges in domains where sample sizes are limited, and statistical inference becomes constrained, with computational speed being a bottleneck in large models of biological systems. As an alternative, we propose the use of conformal inference methods, introducing two novel algorithms that, in some instances, offer non-asymptotic guarantees, enhancing robustness and scalability across various applications. We demonstrate the efficacy of our proposed algorithms through several scenarios, highlighting their advantages over traditional Bayesian approaches. The proposed methods show promising results for diverse biological data structures and scenarios, offering a general framework to quantify uncertainty for dynamic models of biological systems.The software for the methodology and the reproduction of the results is available at https://zenodo.org/doi/10.5281/zenodo.13644870.
Uncertainty quantification in metric spaces
May 08, 2024Abstract:This paper introduces a novel uncertainty quantification framework for regression models where the response takes values in a separable metric space, and the predictors are in a Euclidean space. The proposed algorithms can efficiently handle large datasets and are agnostic to the predictive base model used. Furthermore, the algorithms possess asymptotic consistency guarantees and, in some special homoscedastic cases, we provide non-asymptotic guarantees. To illustrate the effectiveness of the proposed uncertainty quantification framework, we use a linear regression model for metric responses (known as the global Fr\'echet model) in various clinical applications related to precision and digital medicine. The different clinical outcomes analyzed are represented as complex statistical objects, including multivariate Euclidean data, Laplacian graphs, and probability distributions.
Deep Learning Framework with Uncertainty Quantification for Survey Data: Assessing and Predicting Diabetes Mellitus Risk in the American Population
Mar 28, 2024Abstract:Complex survey designs are commonly employed in many medical cohorts. In such scenarios, developing case-specific predictive risk score models that reflect the unique characteristics of the study design is essential. This approach is key to minimizing potential selective biases in results. The objectives of this paper are: (i) To propose a general predictive framework for regression and classification using neural network (NN) modeling, which incorporates survey weights into the estimation process; (ii) To introduce an uncertainty quantification algorithm for model prediction, tailored for data from complex survey designs; (iii) To apply this method in developing robust risk score models to assess the risk of Diabetes Mellitus in the US population, utilizing data from the NHANES 2011-2014 cohort. The theoretical properties of our estimators are designed to ensure minimal bias and the statistical consistency, thereby ensuring that our models yield reliable predictions and contribute novel scientific insights in diabetes research. While focused on diabetes, this NN predictive framework is adaptable to create clinical models for a diverse range of diseases and medical cohorts. The software and the data used in this paper is publicly available on GitHub.
kNN Algorithm for Conditional Mean and Variance Estimation with Automated Uncertainty Quantification and Variable Selection
Feb 02, 2024Abstract:In this paper, we introduce a kNN-based regression method that synergizes the scalability and adaptability of traditional non-parametric kNN models with a novel variable selection technique. This method focuses on accurately estimating the conditional mean and variance of random response variables, thereby effectively characterizing conditional distributions across diverse scenarios.Our approach incorporates a robust uncertainty quantification mechanism, leveraging our prior estimation work on conditional mean and variance. The employment of kNN ensures scalable computational efficiency in predicting intervals and statistical accuracy in line with optimal non-parametric rates. Additionally, we introduce a new kNN semi-parametric algorithm for estimating ROC curves, accounting for covariates. For selecting the smoothing parameter k, we propose an algorithm with theoretical guarantees.Incorporation of variable selection enhances the performance of the method significantly over conventional kNN techniques in various modeling tasks. We validate the approach through simulations in low, moderate, and high-dimensional covariate spaces. The algorithm's effectiveness is particularly notable in biomedical applications as demonstrated in two case studies. Concluding with a theoretical analysis, we highlight the consistency and convergence rate of our method over traditional kNN models, particularly when the underlying regression model takes values in a low-dimensional space.
Kernel Biclustering algorithm in Hilbert Spaces
Aug 07, 2022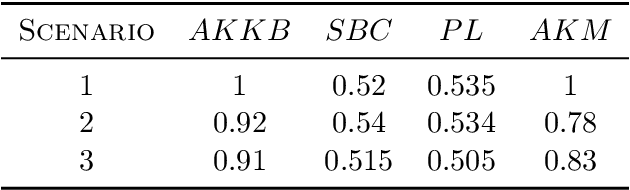
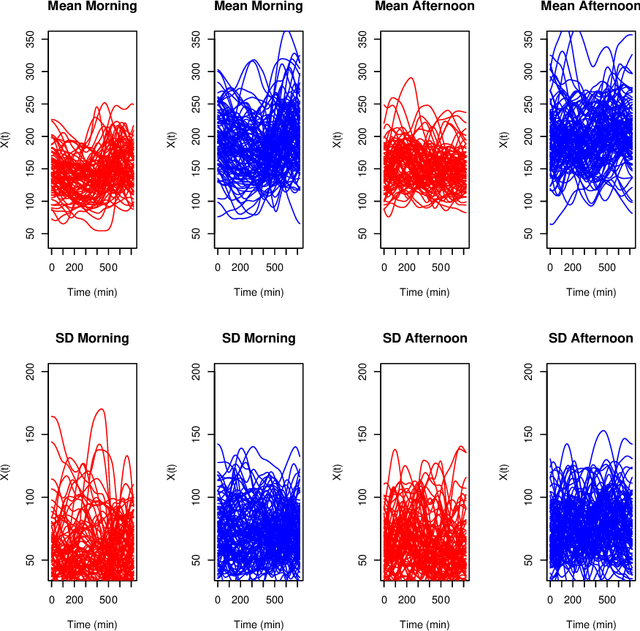
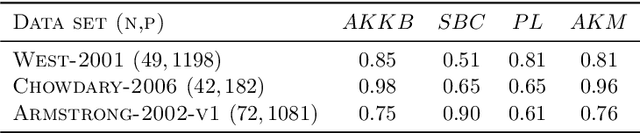
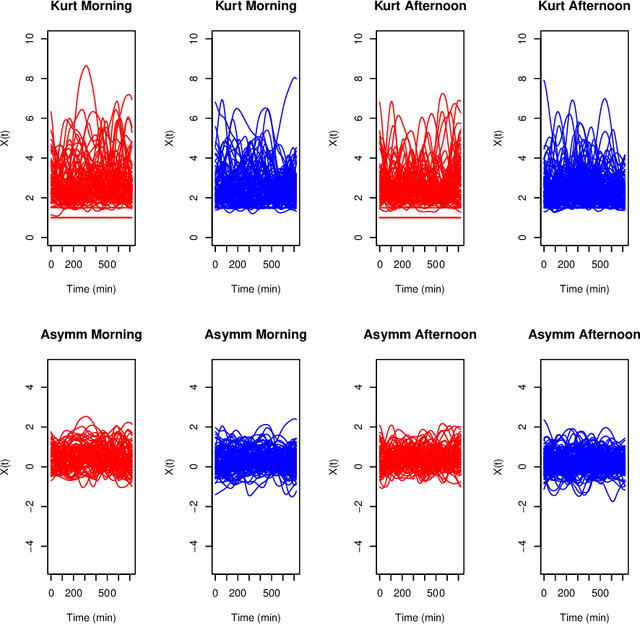
Abstract:Biclustering algorithms partition data and covariates simultaneously, providing new insights in several domains, such as analyzing gene expression to discover new biological functions. This paper develops a new model-free biclustering algorithm in abstract spaces using the notions of energy distance (ED) and the maximum mean discrepancy (MMD) -- two distances between probability distributions capable of handling complex data such as curves or graphs. The proposed method can learn more general and complex cluster shapes than most existing literature approaches, which usually focus on detecting mean and variance differences. Although the biclustering configurations of our approach are constrained to create disjoint structures at the datum and covariate levels, the results are competitive. Our results are similar to state-of-the-art methods in their optimal scenarios, assuming a proper kernel choice, outperforming them when cluster differences are concentrated in higher-order moments. The model's performance has been tested in several situations that involve simulated and real-world datasets. Finally, new theoretical consistency results are established using some tools of the theory of optimal transport.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge