Mara Graziani
We Need Improved Data Curation and Attribution in AI for Scientific Discovery
Apr 03, 2025



Abstract:As the interplay between human-generated and synthetic data evolves, new challenges arise in scientific discovery concerning the integrity of the data and the stability of the models. In this work, we examine the role of synthetic data as opposed to that of real experimental data for scientific research. Our analyses indicate that nearly three-quarters of experimental datasets available on open-access platforms have relatively low adoption rates, opening new opportunities to enhance their discoverability and usability by automated methods. Additionally, we observe an increasing difficulty in distinguishing synthetic from real experimental data. We propose supplementing ongoing efforts in automating synthetic data detection by increasing the focus on watermarking real experimental data, thereby strengthening data traceability and integrity. Our estimates suggest that watermarking even less than half of the real world data generated annually could help sustain model robustness, while promoting a balanced integration of synthetic and human-generated content.
Structural-Based Uncertainty in Deep Learning Across Anatomical Scales: Analysis in White Matter Lesion Segmentation
Nov 15, 2023



Abstract:This paper explores uncertainty quantification (UQ) as an indicator of the trustworthiness of automated deep-learning (DL) tools in the context of white matter lesion (WML) segmentation from magnetic resonance imaging (MRI) scans of multiple sclerosis (MS) patients. Our study focuses on two principal aspects of uncertainty in structured output segmentation tasks. Firstly, we postulate that a good uncertainty measure should indicate predictions likely to be incorrect with high uncertainty values. Second, we investigate the merit of quantifying uncertainty at different anatomical scales (voxel, lesion, or patient). We hypothesize that uncertainty at each scale is related to specific types of errors. Our study aims to confirm this relationship by conducting separate analyses for in-domain and out-of-domain settings. Our primary methodological contributions are (i) the development of novel measures for quantifying uncertainty at lesion and patient scales, derived from structural prediction discrepancies, and (ii) the extension of an error retention curve analysis framework to facilitate the evaluation of UQ performance at both lesion and patient scales. The results from a multi-centric MRI dataset of 172 patients demonstrate that our proposed measures more effectively capture model errors at the lesion and patient scales compared to measures that average voxel-scale uncertainty values. We provide the UQ protocols code at https://github.com/Medical-Image-Analysis-Laboratory/MS_WML_uncs.
Uncovering Unique Concept Vectors through Latent Space Decomposition
Jul 14, 2023Abstract:Interpreting the inner workings of deep learning models is crucial for establishing trust and ensuring model safety. Concept-based explanations have emerged as a superior approach that is more interpretable than feature attribution estimates such as pixel saliency. However, defining the concepts for the interpretability analysis biases the explanations by the user's expectations on the concepts. To address this, we propose a novel post-hoc unsupervised method that automatically uncovers the concepts learned by deep models during training. By decomposing the latent space of a layer in singular vectors and refining them by unsupervised clustering, we uncover concept vectors aligned with directions of high variance that are relevant to the model prediction, and that point to semantically distinct concepts. Our extensive experiments reveal that the majority of our concepts are readily understandable to humans, exhibit coherency, and bear relevance to the task at hand. Moreover, we showcase the practical utility of our method in dataset exploration, where our concept vectors successfully identify outlier training samples affected by various confounding factors. This novel exploration technique has remarkable versatility to data types and model architectures and it will facilitate the identification of biases and the discovery of sources of error within training data.
Disentangling Neuron Representations with Concept Vectors
Apr 19, 2023



Abstract:Mechanistic interpretability aims to understand how models store representations by breaking down neural networks into interpretable units. However, the occurrence of polysemantic neurons, or neurons that respond to multiple unrelated features, makes interpreting individual neurons challenging. This has led to the search for meaningful vectors, known as concept vectors, in activation space instead of individual neurons. The main contribution of this paper is a method to disentangle polysemantic neurons into concept vectors encapsulating distinct features. Our method can search for fine-grained concepts according to the user's desired level of concept separation. The analysis shows that polysemantic neurons can be disentangled into directions consisting of linear combinations of neurons. Our evaluations show that the concept vectors found encode coherent, human-understandable features.
Regression-based Deep-Learning predicts molecular biomarkers from pathology slides
Apr 11, 2023Abstract:Deep Learning (DL) can predict biomarkers from cancer histopathology. Several clinically approved applications use this technology. Most approaches, however, predict categorical labels, whereas biomarkers are often continuous measurements. We hypothesized that regression-based DL outperforms classification-based DL. Therefore, we developed and evaluated a new self-supervised attention-based weakly supervised regression method that predicts continuous biomarkers directly from images in 11,671 patients across nine cancer types. We tested our method for multiple clinically and biologically relevant biomarkers: homologous repair deficiency (HRD) score, a clinically used pan-cancer biomarker, as well as markers of key biological processes in the tumor microenvironment. Using regression significantly enhances the accuracy of biomarker prediction, while also improving the interpretability of the results over classification. In a large cohort of colorectal cancer patients, regression-based prediction scores provide a higher prognostic value than classification-based scores. Our open-source regression approach offers a promising alternative for continuous biomarker analysis in computational pathology.
Tackling Bias in the Dice Similarity Coefficient: Introducing nDSC for White Matter Lesion Segmentation
Feb 10, 2023Abstract:The development of automatic segmentation techniques for medical imaging tasks requires assessment metrics to fairly judge and rank such approaches on benchmarks. The Dice Similarity Coefficient (DSC) is a popular choice for comparing the agreement between the predicted segmentation against a ground-truth mask. However, the DSC metric has been shown to be biased to the occurrence rate of the positive class in the ground-truth, and hence should be considered in combination with other metrics. This work describes a detailed analysis of the recently proposed normalised Dice Similarity Coefficient (nDSC) for binary segmentation tasks as an adaptation of DSC which scales the precision at a fixed recall rate to tackle this bias. White matter lesion segmentation on magnetic resonance images of multiple sclerosis patients is selected as a case study task to empirically assess the suitability of nDSC. We validate the normalised DSC using two different models across 59 subject scans with a wide range of lesion loads. It is found that the nDSC is less biased than DSC with lesion load on standard white matter lesion segmentation benchmarks measured using standard rank correlation coefficients. An implementation of nDSC is made available at: https://github.com/NataliiaMolch/nDSC .
Novel structural-scale uncertainty measures and error retention curves: application to multiple sclerosis
Nov 11, 2022Abstract:This paper focuses on the uncertainty estimation for white matter lesions (WML) segmentation in magnetic resonance imaging (MRI). On one side, voxel-scale segmentation errors cause the erroneous delineation of the lesions; on the other side, lesion-scale detection errors lead to wrong lesion counts. Both of these factors are clinically relevant for the assessment of multiple sclerosis patients. This work aims to compare the ability of different voxel- and lesion-scale uncertainty measures to capture errors related to segmentation and lesion detection, respectively. Our main contributions are (i) proposing new measures of lesion-scale uncertainty that do not utilise voxel-scale uncertainties; (ii) extending an error retention curves analysis framework for evaluation of lesion-scale uncertainty measures. Our results obtained on the multi-center testing set of 58 patients demonstrate that the proposed lesion-scale measure achieves the best performance among the analysed measures. All code implementations are provided at https://github.com/NataliiaMolch/MS_WML_uncs
Attention-based Interpretable Regression of Gene Expression in Histology
Aug 29, 2022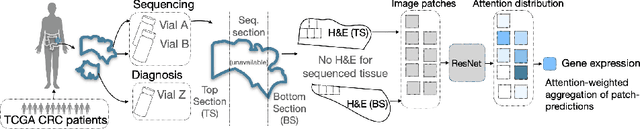
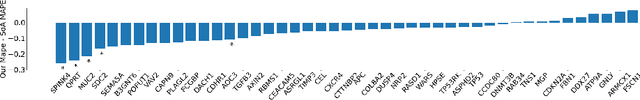
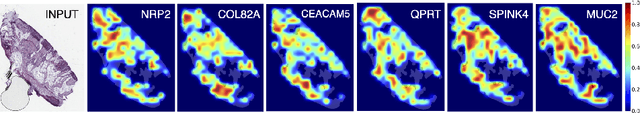

Abstract:Interpretability of deep learning is widely used to evaluate the reliability of medical imaging models and reduce the risks of inaccurate patient recommendations. For models exceeding human performance, e.g. predicting RNA structure from microscopy images, interpretable modelling can be further used to uncover highly non-trivial patterns which are otherwise imperceptible to the human eye. We show that interpretability can reveal connections between the microscopic appearance of cancer tissue and its gene expression profiling. While exhaustive profiling of all genes from the histology images is still challenging, we estimate the expression values of a well-known subset of genes that is indicative of cancer molecular subtype, survival, and treatment response in colorectal cancer. Our approach successfully identifies meaningful information from the image slides, highlighting hotspots of high gene expression. Our method can help characterise how gene expression shapes tissue morphology and this may be beneficial for patient stratification in the pathology unit. The code is available on GitHub.
Shifts 2.0: Extending The Dataset of Real Distributional Shifts
Jun 30, 2022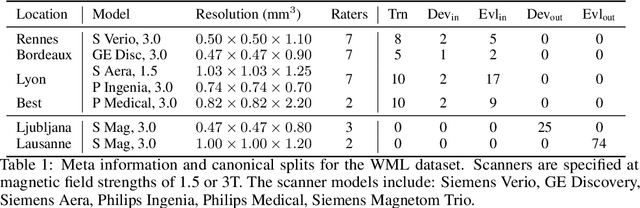
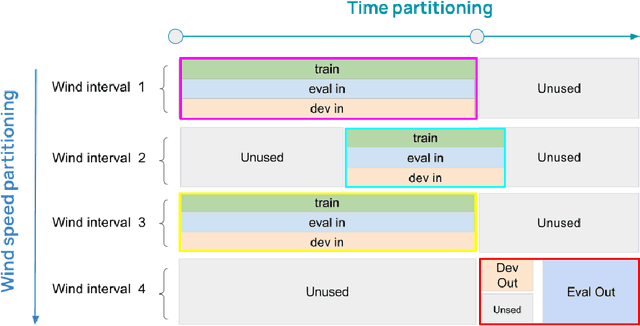
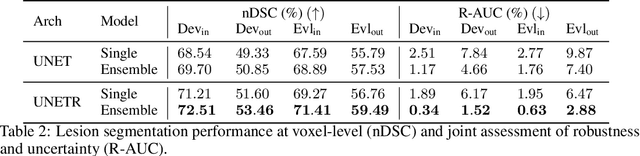
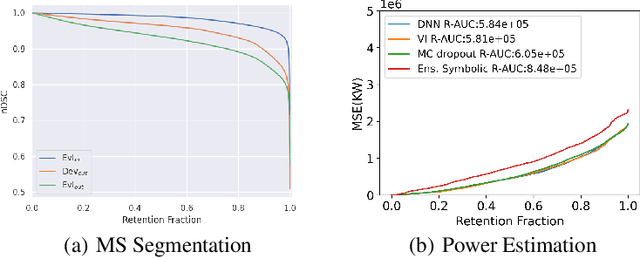
Abstract:Distributional shift, or the mismatch between training and deployment data, is a significant obstacle to the usage of machine learning in high-stakes industrial applications, such as autonomous driving and medicine. This creates a need to be able to assess how robustly ML models generalize as well as the quality of their uncertainty estimates. Standard ML baseline datasets do not allow these properties to be assessed, as the training, validation and test data are often identically distributed. Recently, a range of dedicated benchmarks have appeared, featuring both distributionally matched and shifted data. Among these benchmarks, the Shifts dataset stands out in terms of the diversity of tasks as well as the data modalities it features. While most of the benchmarks are heavily dominated by 2D image classification tasks, Shifts contains tabular weather forecasting, machine translation, and vehicle motion prediction tasks. This enables the robustness properties of models to be assessed on a diverse set of industrial-scale tasks and either universal or directly applicable task-specific conclusions to be reached. In this paper, we extend the Shifts Dataset with two datasets sourced from industrial, high-risk applications of high societal importance. Specifically, we consider the tasks of segmentation of white matter Multiple Sclerosis lesions in 3D magnetic resonance brain images and the estimation of power consumption in marine cargo vessels. Both tasks feature ubiquitous distributional shifts and a strict safety requirement due to the high cost of errors. These new datasets will allow researchers to further explore robust generalization and uncertainty estimation in new situations. In this work, we provide a description of the dataset and baseline results for both tasks.
Guiding CNNs towards Relevant Concepts by Multi-task and Adversarial Learning
Aug 04, 2020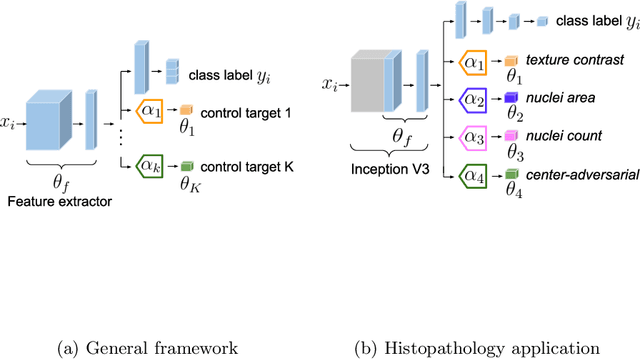
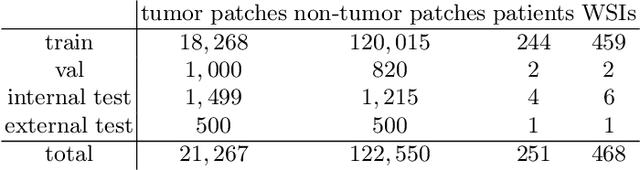
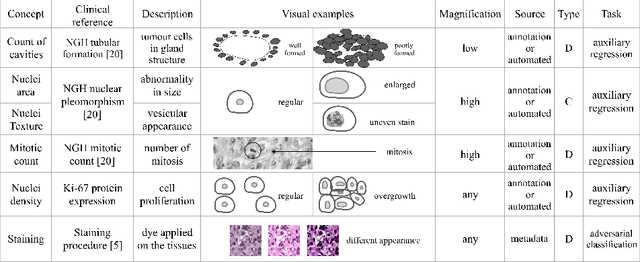
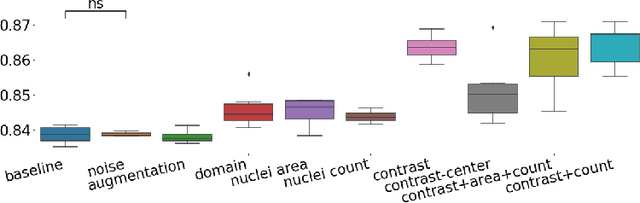
Abstract:The opaqueness of deep learning limits its deployment in critical application scenarios such as cancer grading in medical images. In this paper, a framework for guiding CNN training is built on top of successful existing techniques of hard parameter sharing, with the main goal of explicitly introducing expert knowledge in the training objectives. The learning process is guided by identifying concepts that are relevant or misleading for the task. Relevant concepts are encouraged to appear in the representation through multi-task learning. Undesired and misleading concepts are discouraged by a gradient reversal operation. In this way, a shift in the deep representations can be corrected to match the clinicians' assumptions. The application on breast lymph nodes histopathology data from the Camelyon challenge shows a significant increase in the generalization performance on unseen patients (from 0.839 to 0.864 average AUC, $\text{p-value} = 0,0002$) when the internal representations are controlled by prior knowledge regarding the acquisition center and visual features of the tissue. The code will be shared for reproducibility on our GitHub repository.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge