Jonathan Deaton
Big Self-Supervised Models Advance Medical Image Classification
Jan 13, 2021
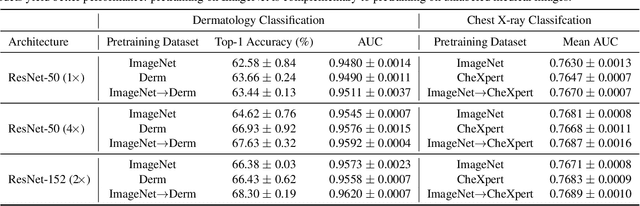
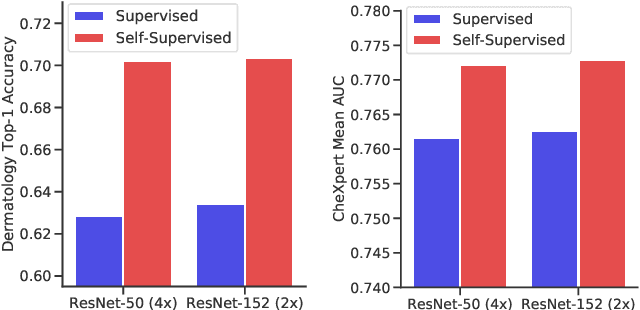
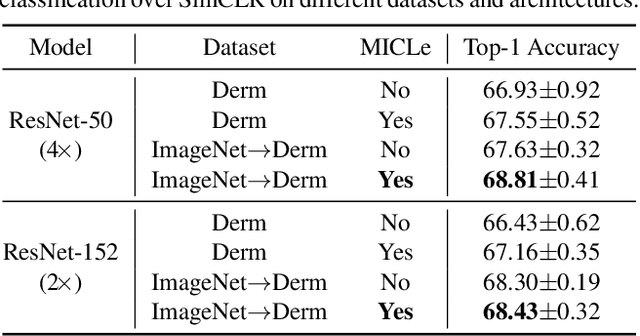
Abstract:Self-supervised pretraining followed by supervised fine-tuning has seen success in image recognition, especially when labeled examples are scarce, but has received limited attention in medical image analysis. This paper studies the effectiveness of self-supervised learning as a pretraining strategy for medical image classification. We conduct experiments on two distinct tasks: dermatology skin condition classification from digital camera images and multi-label chest X-ray classification, and demonstrate that self-supervised learning on ImageNet, followed by additional self-supervised learning on unlabeled domain-specific medical images significantly improves the accuracy of medical image classifiers. We introduce a novel Multi-Instance Contrastive Learning (MICLe) method that uses multiple images of the underlying pathology per patient case, when available, to construct more informative positive pairs for self-supervised learning. Combining our contributions, we achieve an improvement of 6.7% in top-1 accuracy and an improvement of 1.1% in mean AUC on dermatology and chest X-ray classification respectively, outperforming strong supervised baselines pretrained on ImageNet. In addition, we show that big self-supervised models are robust to distribution shift and can learn efficiently with a small number of labeled medical images.
Underspecification Presents Challenges for Credibility in Modern Machine Learning
Nov 06, 2020


Abstract:ML models often exhibit unexpectedly poor behavior when they are deployed in real-world domains. We identify underspecification as a key reason for these failures. An ML pipeline is underspecified when it can return many predictors with equivalently strong held-out performance in the training domain. Underspecification is common in modern ML pipelines, such as those based on deep learning. Predictors returned by underspecified pipelines are often treated as equivalent based on their training domain performance, but we show here that such predictors can behave very differently in deployment domains. This ambiguity can lead to instability and poor model behavior in practice, and is a distinct failure mode from previously identified issues arising from structural mismatch between training and deployment domains. We show that this problem appears in a wide variety of practical ML pipelines, using examples from computer vision, medical imaging, natural language processing, clinical risk prediction based on electronic health records, and medical genomics. Our results show the need to explicitly account for underspecification in modeling pipelines that are intended for real-world deployment in any domain.
Addressing the Real-world Class Imbalance Problem in Dermatology
Oct 09, 2020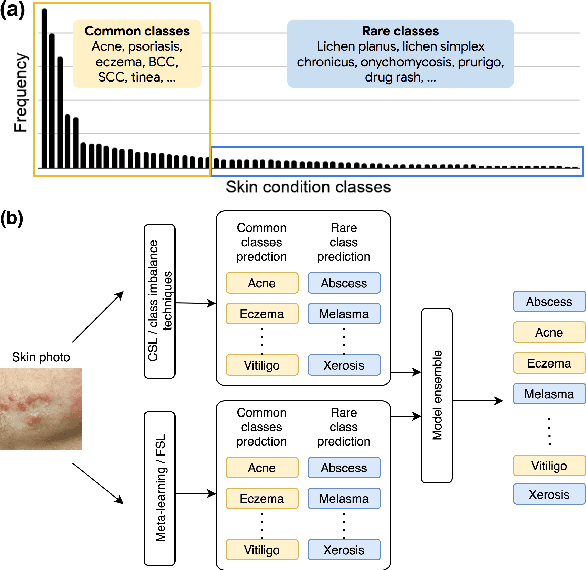
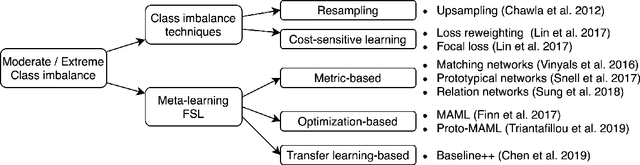
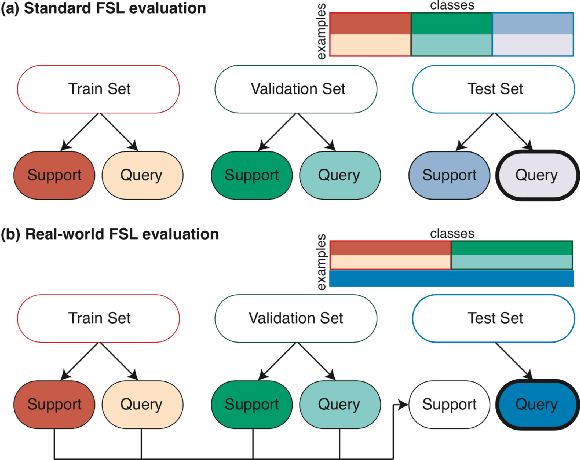
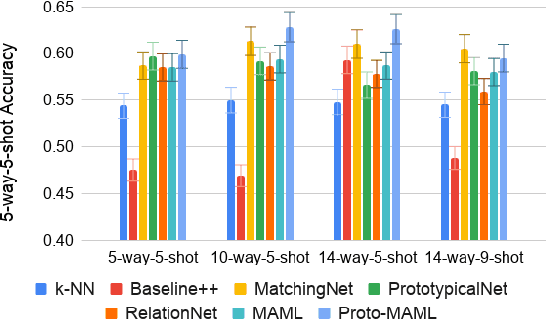
Abstract:Class imbalance is a common problem in medical diagnosis, causing a standard classifier to be biased towards the common classes and perform poorly on the rare classes. This is especially true for dermatology, a specialty with thousands of skin conditions but many of which have rare prevalence in the real world. Motivated by recent advances, we explore few-shot learning methods as well as conventional class imbalance techniques for the skin condition recognition problem and propose an evaluation setup to fairly assess the real-world utility of such approaches. When compared to conventional class imbalance techniques, we find that few-shot learning methods are not as performant as those conventional methods, but combining the two approaches using a novel ensemble leads to improvement in model performance, especially for rare classes. We conclude that the ensemble can be useful to address the class imbalance problem, yet progress here can further be accelerated by the use of real-world evaluation setups for benchmarking new methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge