Joao Bettencourt-Silva
Query-driven Document-level Scientific Evidence Extraction from Biomedical Studies
May 09, 2025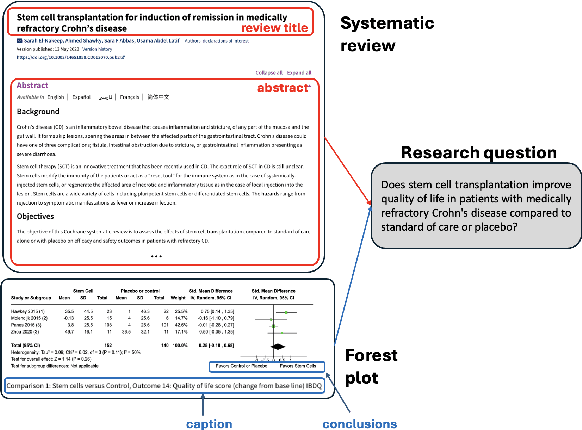
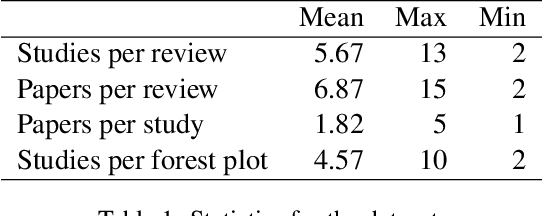
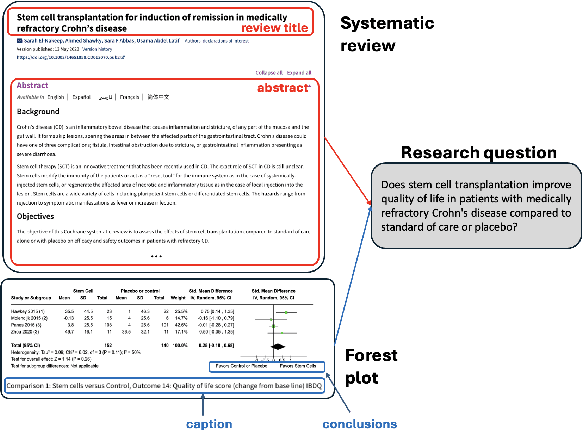
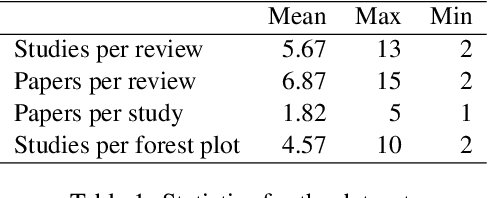
Abstract:Extracting scientific evidence from biomedical studies for clinical research questions (e.g., Does stem cell transplantation improve quality of life in patients with medically refractory Crohn's disease compared to placebo?) is a crucial step in synthesising biomedical evidence. In this paper, we focus on the task of document-level scientific evidence extraction for clinical questions with conflicting evidence. To support this task, we create a dataset called CochraneForest, leveraging forest plots from Cochrane systematic reviews. It comprises 202 annotated forest plots, associated clinical research questions, full texts of studies, and study-specific conclusions. Building on CochraneForest, we propose URCA (Uniform Retrieval Clustered Augmentation), a retrieval-augmented generation framework designed to tackle the unique challenges of evidence extraction. Our experiments show that URCA outperforms the best existing methods by up to 10.3% in F1 score on this task. However, the results also underscore the complexity of CochraneForest, establishing it as a challenging testbed for advancing automated evidence synthesis systems.
Evaluating the Predictive Features of Person-Centric Knowledge Graph Embeddings: Unfolding Ablation Studies
Aug 29, 2024

Abstract:Developing novel predictive models with complex biomedical information is challenging due to various idiosyncrasies related to heterogeneity, standardization or sparseness of the data. We previously introduced a person-centric ontology to organize information about individual patients, and a representation learning framework to extract person-centric knowledge graphs (PKGs) and to train Graph Neural Networks (GNNs). In this paper, we propose a systematic approach to examine the results of GNN models trained with both structured and unstructured information from the MIMIC-III dataset. Through ablation studies on different clinical, demographic, and social data, we show the robustness of this approach in identifying predictive features in PKGs for the task of readmission prediction.
* Published in the 34th Medical Informatics Europe Conference
Representation Learning for Person or Entity-centric Knowledge Graphs: An Application in Healthcare
May 10, 2023



Abstract:Knowledge graphs (KGs) are a popular way to organise information based on ontologies or schemas and have been used across a variety of scenarios from search to recommendation. Despite advances in KGs, representing knowledge remains a non-trivial task across industries and it is especially challenging in the biomedical and healthcare domains due to complex interdependent relations between entities, heterogeneity, lack of standardization, and sparseness of data. KGs are used to discover diagnoses or prioritize genes relevant to disease, but they often rely on schemas that are not centred around a node or entity of interest, such as a person. Entity-centric KGs are relatively unexplored but hold promise in representing important facets connected to a central node and unlocking downstream tasks beyond graph traversal and reasoning, such as generating graph embeddings and training graph neural networks for a wide range of predictive tasks. This paper presents an end-to-end representation learning framework to extract entity-centric KGs from structured and unstructured data. We introduce a star-shaped ontology to represent the multiple facets of a person and use it to guide KG creation. Compact representations of the graphs are created leveraging graph neural networks and experiments are conducted using different levels of heterogeneity or explicitness. A readmission prediction task is used to evaluate the results of the proposed framework, showing a stable system, robust to missing data, that outperforms a range of baseline machine learning classifiers. We highlight that this approach has several potential applications across domains and is open-sourced. Lastly, we discuss lessons learned, challenges, and next steps for the adoption of the framework in practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge