Joana B. Pereira
Global graph features unveiled by unsupervised geometric deep learning
Mar 07, 2025



Abstract:Graphs provide a powerful framework for modeling complex systems, but their structural variability makes analysis and classification challenging. To address this, we introduce GAUDI (Graph Autoencoder Uncovering Descriptive Information), a novel unsupervised geometric deep learning framework that captures both local details and global structure. GAUDI employs an innovative hourglass architecture with hierarchical pooling and upsampling layers, linked through skip connections to preserve essential connectivity information throughout the encoding-decoding process. By mapping different realizations of a system - generated from the same underlying parameters - into a continuous, structured latent space, GAUDI disentangles invariant process-level features from stochastic noise. We demonstrate its power across multiple applications, including modeling small-world networks, characterizing protein assemblies from super-resolution microscopy, analyzing collective motion in the Vicsek model, and capturing age-related changes in brain connectivity. This approach not only improves the analysis of complex graphs but also provides new insights into emergent phenomena across diverse scientific domains.
Roadmap on Deep Learning for Microscopy
Mar 07, 2023



Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Neural Network Training with Highly Incomplete Datasets
Jul 01, 2021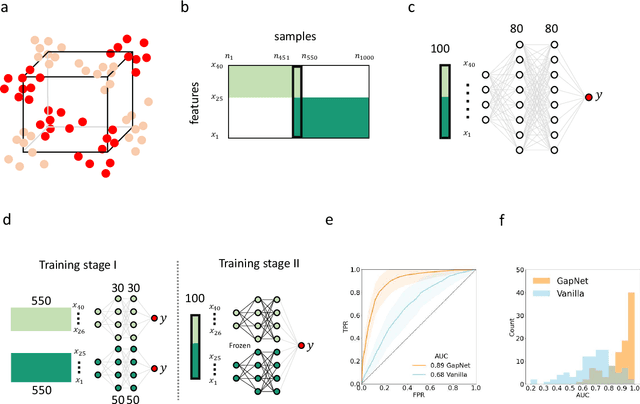
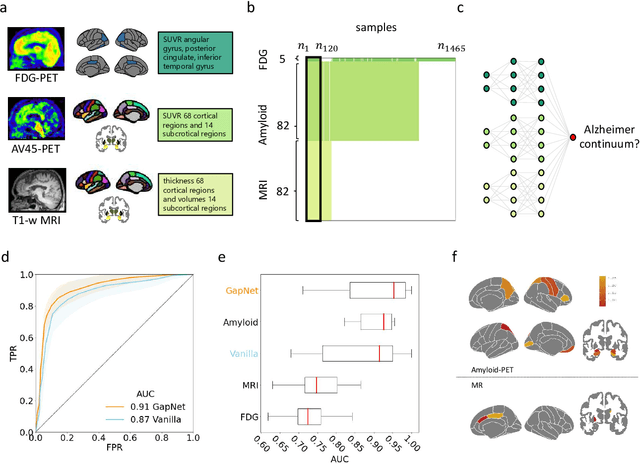
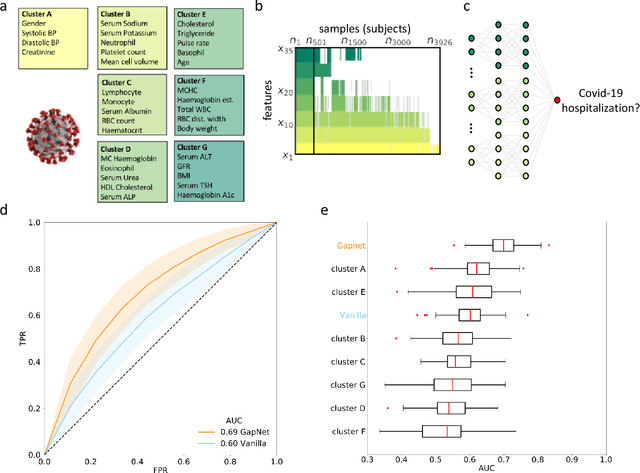
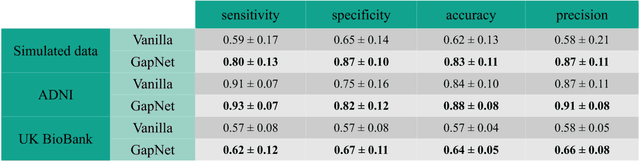
Abstract:Neural network training and validation rely on the availability of large high-quality datasets. However, in many cases only incomplete datasets are available, particularly in health care applications, where each patient typically undergoes different clinical procedures or can drop out of a study. Since the data to train the neural networks need to be complete, most studies discard the incomplete datapoints, which reduces the size of the training data, or impute the missing features, which can lead to artefacts. Alas, both approaches are inadequate when a large portion of the data is missing. Here, we introduce GapNet, an alternative deep-learning training approach that can use highly incomplete datasets. First, the dataset is split into subsets of samples containing all values for a certain cluster of features. Then, these subsets are used to train individual neural networks. Finally, this ensemble of neural networks is combined into a single neural network whose training is fine-tuned using all complete datapoints. Using two highly incomplete real-world medical datasets, we show that GapNet improves the identification of patients with underlying Alzheimer's disease pathology and of patients at risk of hospitalization due to Covid-19. By distilling the information available in incomplete datasets without having to reduce their size or to impute missing values, GapNet will permit to extract valuable information from a wide range of datasets, benefiting diverse fields from medicine to engineering.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge