Gilles Vandewiele
REFORMS: Reporting Standards for Machine Learning Based Science
Aug 15, 2023

Abstract:Machine learning (ML) methods are proliferating in scientific research. However, the adoption of these methods has been accompanied by failures of validity, reproducibility, and generalizability. These failures can hinder scientific progress, lead to false consensus around invalid claims, and undermine the credibility of ML-based science. ML methods are often applied and fail in similar ways across disciplines. Motivated by this observation, our goal is to provide clear reporting standards for ML-based science. Drawing from an extensive review of past literature, we present the REFORMS checklist ($\textbf{Re}$porting Standards $\textbf{For}$ $\textbf{M}$achine Learning Based $\textbf{S}$cience). It consists of 32 questions and a paired set of guidelines. REFORMS was developed based on a consensus of 19 researchers across computer science, data science, mathematics, social sciences, and biomedical sciences. REFORMS can serve as a resource for researchers when designing and implementing a study, for referees when reviewing papers, and for journals when enforcing standards for transparency and reproducibility.
Perfectly predicting ICU length of stay: too good to be true
Nov 10, 2022


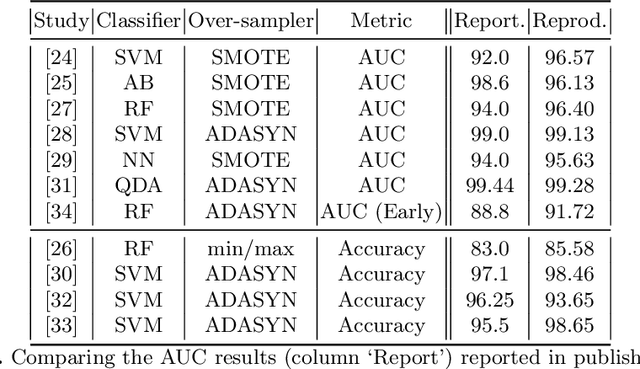
Abstract:A paper of Alsinglawi et al was recently accepted and published in Scientific Reports. In this paper, the authors aim to predict length of stay (LOS), discretized into either long (> 7 days) or short stays (< 7 days), of lung cancer patients in an ICU department using various machine learning techniques. The authors claim to achieve perfect results with an Area Under the Receiver Operating Characteristic curve (AUROC) of 100% with a Random Forest (RF) classifier with ADASYN class balancing over sampling technique, which if accurate could have significant implications for hospital management. However, we have identified several methodological flaws within the manuscript which cause the results to be overly optimistic and would have serious consequences if used in a clinical practice. Moreover, the reporting of the methodology is unclear and many important details are missing from the manuscript, which makes reproduction extremely difficult. We highlight the effect these oversights have had on the result and provide a more believable result of 88.91% AUROC when these oversights are corrected.
Do Not Sleep on Linear Models: Simple and Interpretable Techniques Outperform Deep Learning for Sleep Scoring
Jul 19, 2022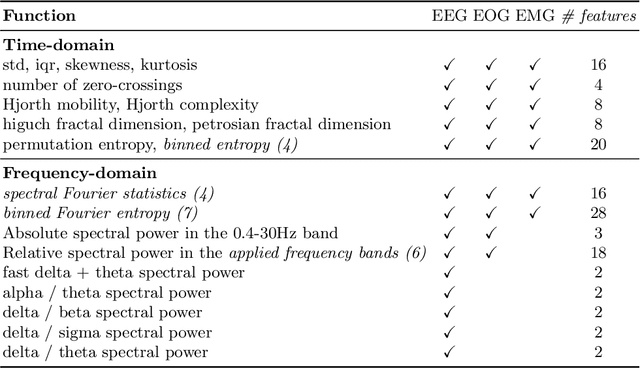
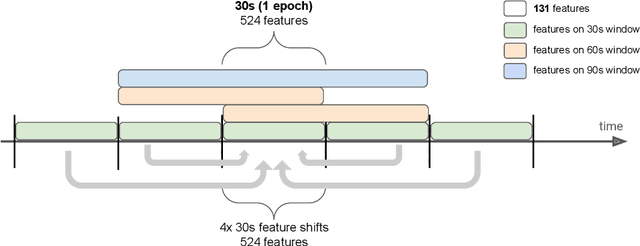
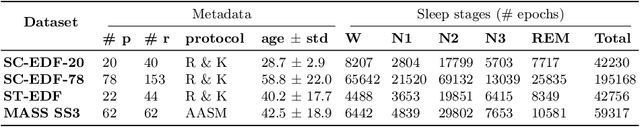
Abstract:Over the last few years, research in automatic sleep scoring has mainly focused on developing increasingly complex deep learning architectures. However, recently these approaches achieved only marginal improvements, often at the expense of requiring more data and more expensive training procedures. Despite all these efforts and their satisfactory performance, automatic sleep staging solutions are not widely adopted in a clinical context yet. We argue that most deep learning solutions for sleep scoring are limited in their real-world applicability as they are hard to train, deploy, and reproduce. Moreover, these solutions lack interpretability and transparency, which are often key to increase adoption rates. In this work, we revisit the problem of sleep stage classification using classical machine learning. Results show that state-of-the-art performance can be achieved with a conventional machine learning pipeline consisting of preprocessing, feature extraction, and a simple machine learning model. In particular, we analyze the performance of a linear model and a non-linear (gradient boosting) model. Our approach surpasses state-of-the-art (that uses the same data) on two public datasets: Sleep-EDF SC-20 (MF1 0.810) and Sleep-EDF ST (MF1 0.795), while achieving competitive results on Sleep-EDF SC-78 (MF1 0.775) and MASS SS3 (MF1 0.817). We show that, for the sleep stage scoring task, the expressiveness of an engineered feature vector is on par with the internally learned representations of deep learning models. This observation opens the door to clinical adoption, as a representative feature vector allows to leverage both the interpretability and successful track record of traditional machine learning models.
pyRDF2Vec: A Python Implementation and Extension of RDF2Vec
May 04, 2022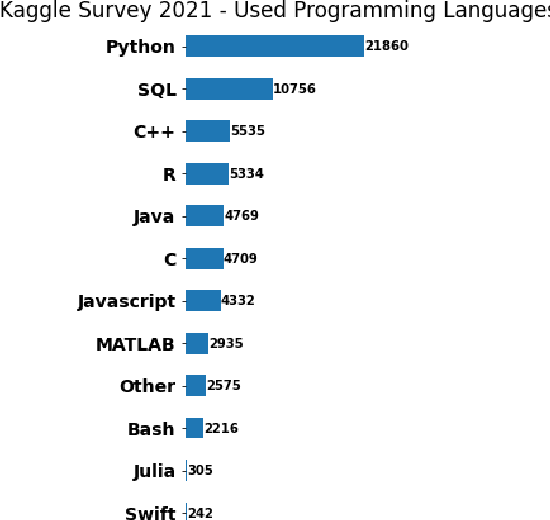
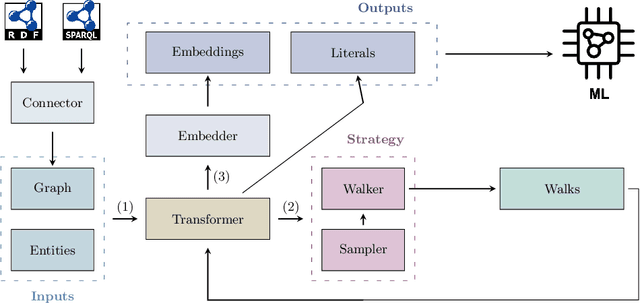
Abstract:This paper introduces pyRDF2Vec, a Python software package that reimplements the well-known RDF2Vec algorithm along with several of its extensions. By making the algorithm available in the most popular data science language, and by bundling all extensions into a single place, the use of RDF2Vec is simplified for data scientists. The package is released under a MIT license and structured in such a way to foster further research into sampling, walking, and embedding strategies, which are vital components of the RDF2Vec algorithm. Several optimisations have been implemented in \texttt{pyRDF2Vec} that allow for more efficient walk extraction than the original algorithm. Furthermore, best practices in terms of code styling, testing, and documentation were applied such that the package is future-proof as well as to facilitate external contributions.
R-GCN: The R Could Stand for Random
Mar 04, 2022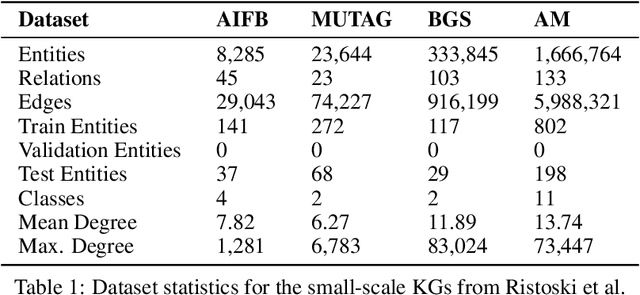
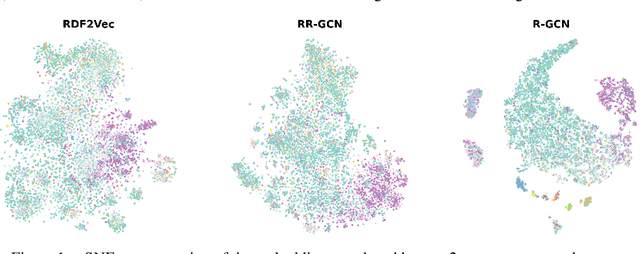
Abstract:The inception of Relational Graph Convolutional Networks (R-GCNs) marked a milestone in the Semantic Web domain as it allows for end-to-end training of machine learning models that operate on Knowledge Graphs (KGs). R-GCNs generate a representation for a node of interest by repeatedly aggregating parametrised, relation-specific transformations of its neighbours. However, in this paper, we argue that the the R-GCN's main contribution lies in this "message passing" paradigm, rather than the learned parameters. To this end, we introduce the "Random Relational Graph Convolutional Network" (RR-GCN), which constructs embeddings for nodes in the KG by aggregating randomly transformed random information from neigbours, i.e., with no learned parameters. We empirically show that RR-GCNs can compete with fully trained R-GCNs in both node classification and link prediction settings. The implications of these results are two-fold: on the one hand, our technique can be used as a quick baseline that novel KG embedding methods should be able to beat. On the other hand, it demonstrates that further research might reveal more parameter-efficient inductive biases for KGs.
Predictive models of RNA degradation through dual crowdsourcing
Oct 14, 2021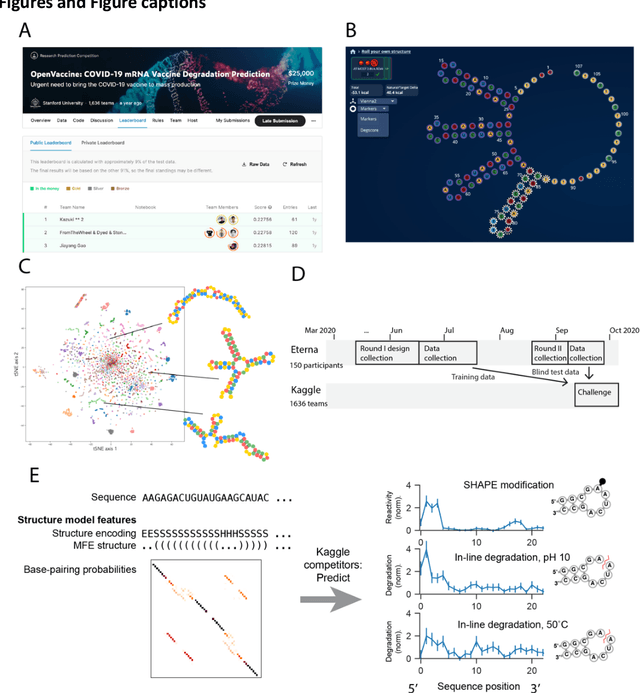
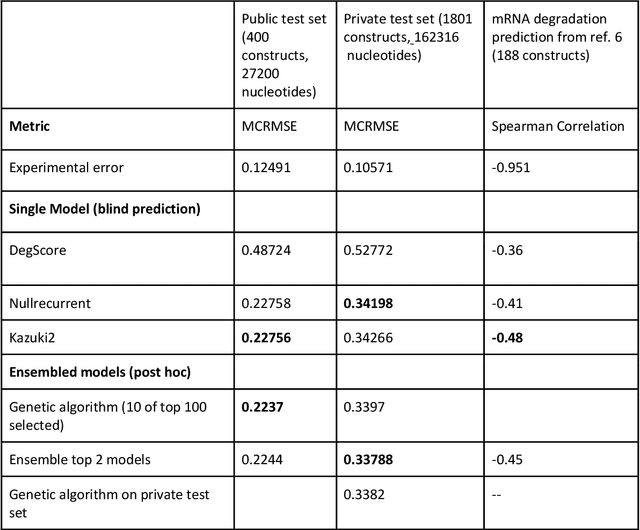
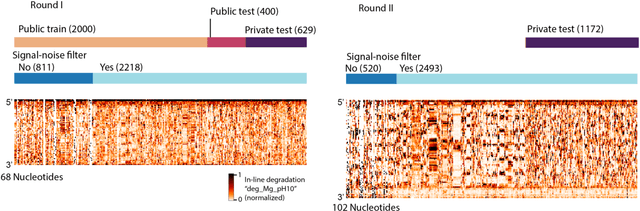
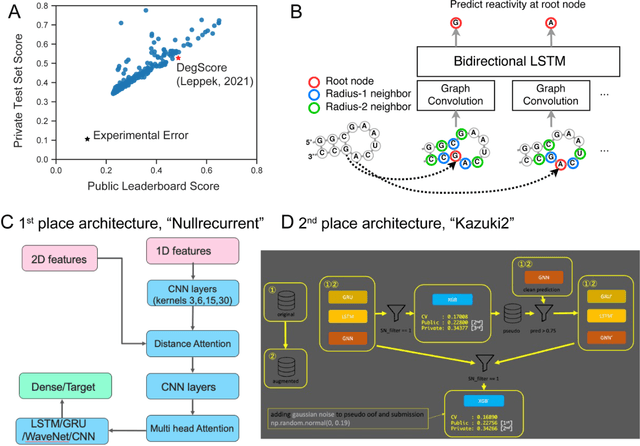
Abstract:Messenger RNA-based medicines hold immense potential, as evidenced by their rapid deployment as COVID-19 vaccines. However, worldwide distribution of mRNA molecules has been limited by their thermostability, which is fundamentally limited by the intrinsic instability of RNA molecules to a chemical degradation reaction called in-line hydrolysis. Predicting the degradation of an RNA molecule is a key task in designing more stable RNA-based therapeutics. Here, we describe a crowdsourced machine learning competition ("Stanford OpenVaccine") on Kaggle, involving single-nucleotide resolution measurements on 6043 102-130-nucleotide diverse RNA constructs that were themselves solicited through crowdsourcing on the RNA design platform Eterna. The entire experiment was completed in less than 6 months. Winning models demonstrated test set errors that were better by 50% than the previous state-of-the-art DegScore model. Furthermore, these models generalized to blindly predicting orthogonal degradation data on much longer mRNA molecules (504-1588 nucleotides) with improved accuracy over DegScore and other models. Top teams integrated natural language processing architectures and data augmentation techniques with predictions from previous dynamic programming models for RNA secondary structure. These results indicate that such models are capable of representing in-line hydrolysis with excellent accuracy, supporting their use for designing stabilized messenger RNAs. The integration of two crowdsourcing platforms, one for data set creation and another for machine learning, may be fruitful for other urgent problems that demand scientific discovery on rapid timescales.
Walk Extraction Strategies for Node Embeddings with RDF2Vec in Knowledge Graphs
Sep 09, 2020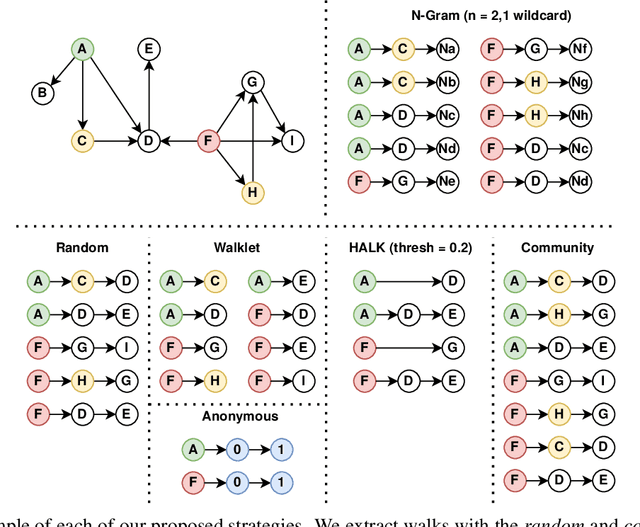
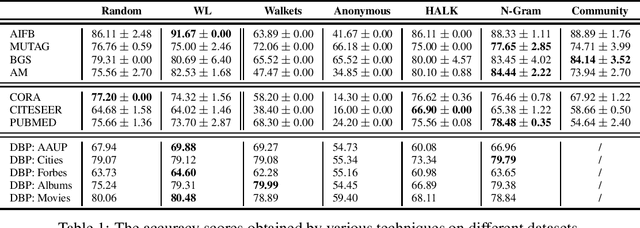
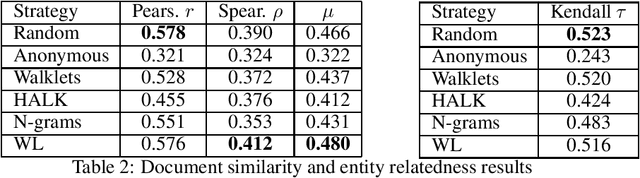
Abstract:As KGs are symbolic constructs, specialized techniques have to be applied in order to make them compatible with data mining techniques. RDF2Vec is an unsupervised technique that can create task-agnostic numerical representations of the nodes in a KG by extending successful language modelling techniques. The original work proposed the Weisfeiler-Lehman (WL) kernel to improve the quality of the representations. However, in this work, we show both formally and empirically that the WL kernel does little to improve walk embeddings in the context of a single KG. As an alternative to the WL kernel, we propose five different strategies to extract information complementary to basic random walks. We compare these walks on several benchmark datasets to show that the \emph{n-gram} strategy performs best on average on node classification tasks and that tuning the walk strategy can result in improved predictive performances.
Overly Optimistic Prediction Results on Imbalanced Data: Flaws and Benefits of Applying Over-sampling
Jan 15, 2020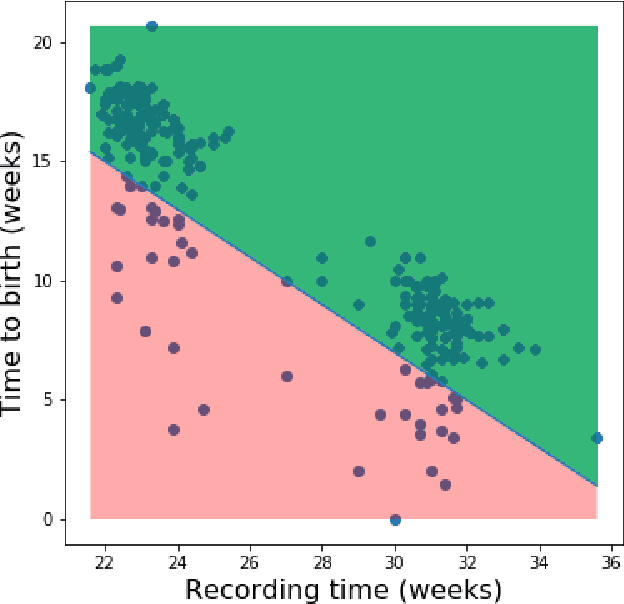
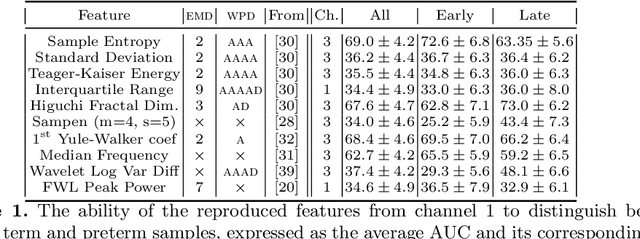
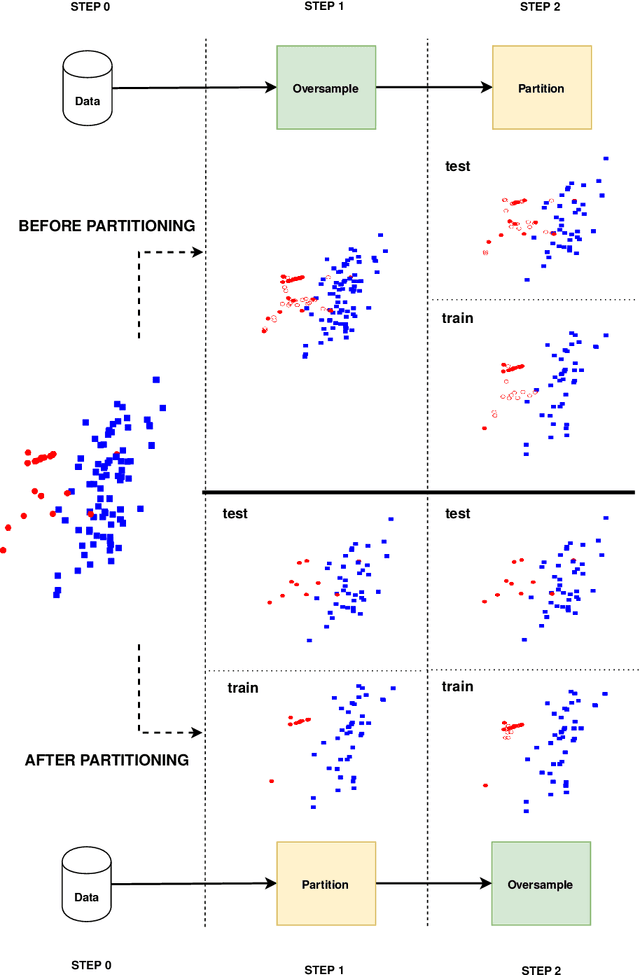
Abstract:Information extracted from electrohysterography recordings could potentially prove to be an interesting additional source of information to estimate the risk on preterm birth. Recently, a large number of studies have reported near-perfect results to distinguish between recordings of patients that will deliver term or preterm using a public resource, called the Term/Preterm Electrohysterogram database. However, we argue that these results are overly optimistic due to a methodological flaw being made. In this work, we focus on one specific type of methodological flaw: applying over-sampling before partitioning the data into mutually exclusive training and testing sets. We show how this causes the results to be biased using two artificial datasets and reproduce results of studies in which this flaw was identified. Moreover, we evaluate the actual impact of over-sampling on predictive performance, when applied prior to data partitioning, using the same methodologies of related studies, to provide a realistic view of these methodologies' generalization capabilities. We make our research reproducible by providing all the code under an open license.
GENESIM: genetic extraction of a single, interpretable model
Nov 17, 2016
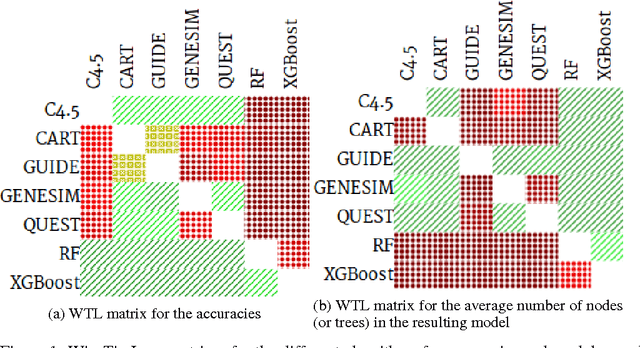
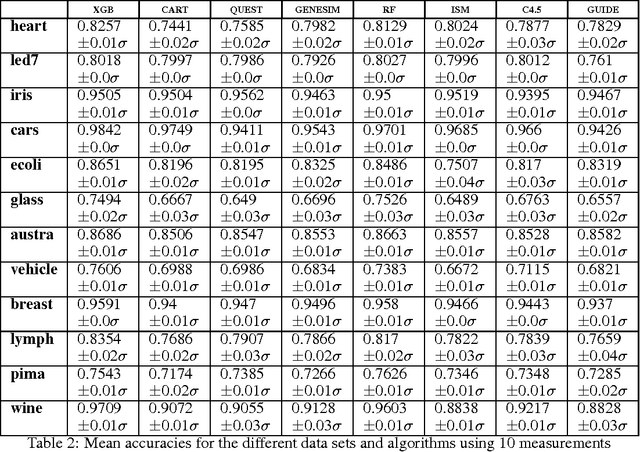
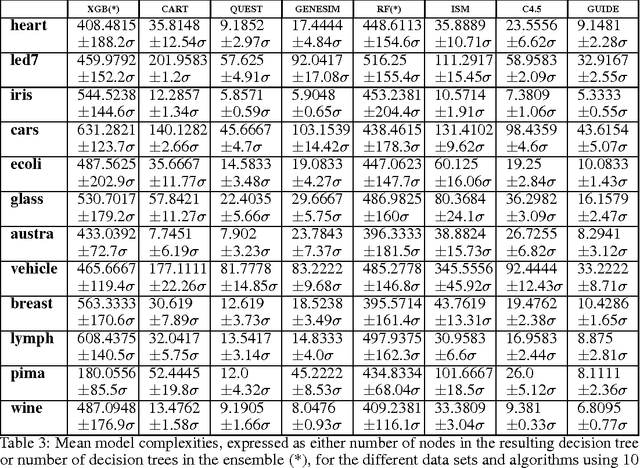
Abstract:Models obtained by decision tree induction techniques excel in being interpretable.However, they can be prone to overfitting, which results in a low predictive performance. Ensemble techniques are able to achieve a higher accuracy. However, this comes at a cost of losing interpretability of the resulting model. This makes ensemble techniques impractical in applications where decision support, instead of decision making, is crucial. To bridge this gap, we present the GENESIM algorithm that transforms an ensemble of decision trees to a single decision tree with an enhanced predictive performance by using a genetic algorithm. We compared GENESIM to prevalent decision tree induction and ensemble techniques using twelve publicly available data sets. The results show that GENESIM achieves a better predictive performance on most of these data sets than decision tree induction techniques and a predictive performance in the same order of magnitude as the ensemble techniques. Moreover, the resulting model of GENESIM has a very low complexity, making it very interpretable, in contrast to ensemble techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge