Georg Widhalm
Automatic and standardized surgical reporting for central nervous system tumors
Aug 12, 2025Abstract:Magnetic resonance (MR) imaging is essential for evaluating central nervous system (CNS) tumors, guiding surgical planning, treatment decisions, and assessing postoperative outcomes and complication risks. While recent work has advanced automated tumor segmentation and report generation, most efforts have focused on preoperative data, with limited attention to postoperative imaging analysis. This study introduces a comprehensive pipeline for standardized postsurtical reporting in CNS tumors. Using the Attention U-Net architecture, segmentation models were trained for the preoperative (non-enhancing) tumor core, postoperative contrast-enhancing residual tumor, and resection cavity. Additionally, MR sequence classification and tumor type identification for contrast-enhancing lesions were explored using the DenseNet architecture. The models were integrated into a reporting pipeline, following the RANO 2.0 guidelines. Training was conducted on multicentric datasets comprising 2000 to 7000 patients, using a 5-fold cross-validation. Evaluation included patient-, voxel-, and object-wise metrics, with benchmarking against the latest BraTS challenge results. The segmentation models achieved average voxel-wise Dice scores of 87%, 66%, 70%, and 77% for the tumor core, non-enhancing tumor core, contrast-enhancing residual tumor, and resection cavity, respectively. Classification models reached 99.5% balanced accuracy in MR sequence classification and 80% in tumor type classification. The pipeline presented in this study enables robust, automated segmentation, MR sequence classification, and standardized report generation aligned with RANO 2.0 guidelines, enhancing postoperative evaluation and clinical decision-making. The proposed models and methods were integrated into Raidionics, open-source software platform for CNS tumor analysis, now including a dedicated module for postsurgical analysis.
A Flow-based Truncated Denoising Diffusion Model for Super-resolution Magnetic Resonance Spectroscopic Imaging
Oct 25, 2024Abstract:Magnetic Resonance Spectroscopic Imaging (MRSI) is a non-invasive imaging technique for studying metabolism and has become a crucial tool for understanding neurological diseases, cancers and diabetes. High spatial resolution MRSI is needed to characterize lesions, but in practice MRSI is acquired at low resolution due to time and sensitivity restrictions caused by the low metabolite concentrations. Therefore, there is an imperative need for a post-processing approach to generate high-resolution MRSI from low-resolution data that can be acquired fast and with high sensitivity. Deep learning-based super-resolution methods provided promising results for improving the spatial resolution of MRSI, but they still have limited capability to generate accurate and high-quality images. Recently, diffusion models have demonstrated superior learning capability than other generative models in various tasks, but sampling from diffusion models requires iterating through a large number of diffusion steps, which is time-consuming. This work introduces a Flow-based Truncated Denoising Diffusion Model (FTDDM) for super-resolution MRSI, which shortens the diffusion process by truncating the diffusion chain, and the truncated steps are estimated using a normalizing flow-based network. The network is conditioned on upscaling factors to enable multi-scale super-resolution. To train and evaluate the deep learning models, we developed a 1H-MRSI dataset acquired from 25 high-grade glioma patients. We demonstrate that FTDDM outperforms existing generative models while speeding up the sampling process by over 9-fold compared to the baseline diffusion model. Neuroradiologists' evaluations confirmed the clinical advantages of our method, which also supports uncertainty estimation and sharpness adjustment, extending its potential clinical applications.
* Accepted by Medical Image Analysis (MedIA)
Segmentation of glioblastomas in early post-operative multi-modal MRI with deep neural networks
Apr 18, 2023Abstract:Extent of resection after surgery is one of the main prognostic factors for patients diagnosed with glioblastoma. To achieve this, accurate segmentation and classification of residual tumor from post-operative MR images is essential. The current standard method for estimating it is subject to high inter- and intra-rater variability, and an automated method for segmentation of residual tumor in early post-operative MRI could lead to a more accurate estimation of extent of resection. In this study, two state-of-the-art neural network architectures for pre-operative segmentation were trained for the task. The models were extensively validated on a multicenter dataset with nearly 1000 patients, from 12 hospitals in Europe and the United States. The best performance achieved was a 61\% Dice score, and the best classification performance was about 80\% balanced accuracy, with a demonstrated ability to generalize across hospitals. In addition, the segmentation performance of the best models was on par with human expert raters. The predicted segmentations can be used to accurately classify the patients into those with residual tumor, and those with gross total resection.
Artificial-intelligence-based molecular classification of diffuse gliomas using rapid, label-free optical imaging
Mar 23, 2023Abstract:Molecular classification has transformed the management of brain tumors by enabling more accurate prognostication and personalized treatment. However, timely molecular diagnostic testing for patients with brain tumors is limited, complicating surgical and adjuvant treatment and obstructing clinical trial enrollment. In this study, we developed DeepGlioma, a rapid ($< 90$ seconds), artificial-intelligence-based diagnostic screening system to streamline the molecular diagnosis of diffuse gliomas. DeepGlioma is trained using a multimodal dataset that includes stimulated Raman histology (SRH); a rapid, label-free, non-consumptive, optical imaging method; and large-scale, public genomic data. In a prospective, multicenter, international testing cohort of patients with diffuse glioma ($n=153$) who underwent real-time SRH imaging, we demonstrate that DeepGlioma can predict the molecular alterations used by the World Health Organization to define the adult-type diffuse glioma taxonomy (IDH mutation, 1p19q co-deletion and ATRX mutation), achieving a mean molecular classification accuracy of $93.3\pm 1.6\%$. Our results represent how artificial intelligence and optical histology can be used to provide a rapid and scalable adjunct to wet lab methods for the molecular screening of patients with diffuse glioma.
Flow-based Visual Quality Enhancer for Super-resolution Magnetic Resonance Spectroscopic Imaging
Jul 20, 2022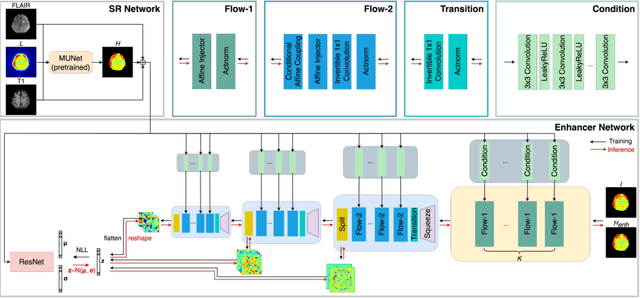
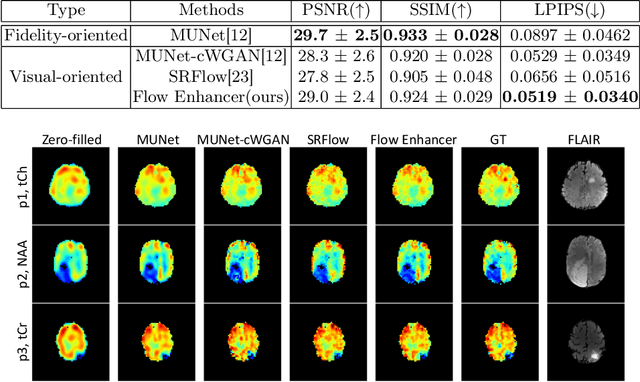
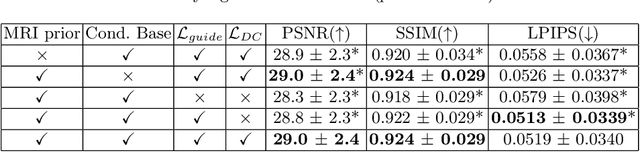
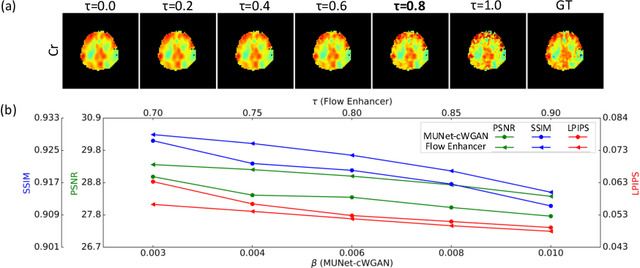
Abstract:Magnetic Resonance Spectroscopic Imaging (MRSI) is an essential tool for quantifying metabolites in the body, but the low spatial resolution limits its clinical applications. Deep learning-based super-resolution methods provided promising results for improving the spatial resolution of MRSI, but the super-resolved images are often blurry compared to the experimentally-acquired high-resolution images. Attempts have been made with the generative adversarial networks to improve the image visual quality. In this work, we consider another type of generative model, the flow-based model, of which the training is more stable and interpretable compared to the adversarial networks. Specifically, we propose a flow-based enhancer network to improve the visual quality of super-resolution MRSI. Different from previous flow-based models, our enhancer network incorporates anatomical information from additional image modalities (MRI) and uses a learnable base distribution. In addition, we impose a guide loss and a data-consistency loss to encourage the network to generate images with high visual quality while maintaining high fidelity. Experiments on a 1H-MRSI dataset acquired from 25 high-grade glioma patients indicate that our enhancer network outperforms the adversarial networks and the baseline flow-based methods. Our method also allows visual quality adjustment and uncertainty estimation.
Multi-scale Super-resolution Magnetic Resonance Spectroscopic Imaging with Adjustable Sharpness
Jun 17, 2022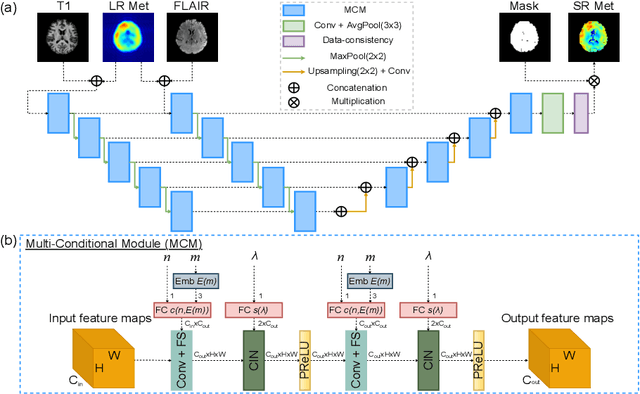
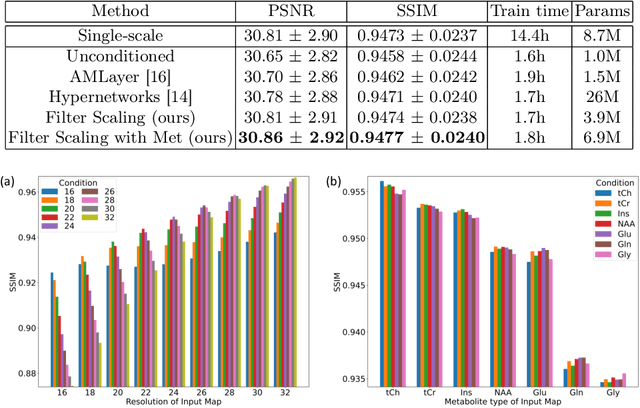
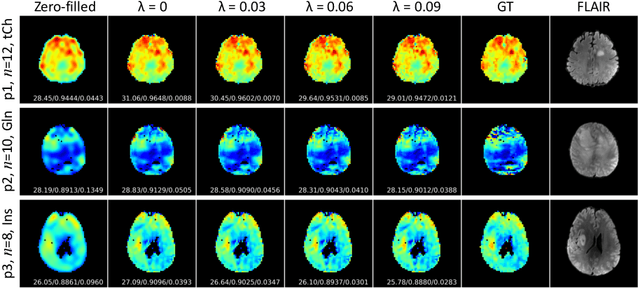
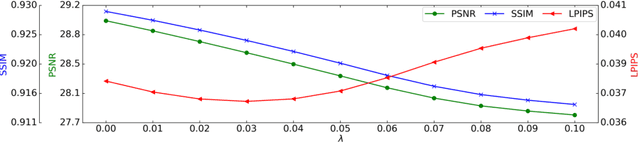
Abstract:Magnetic Resonance Spectroscopic Imaging (MRSI) is a valuable tool for studying metabolic activities in the human body, but the current applications are limited to low spatial resolutions. The existing deep learning-based MRSI super-resolution methods require training a separate network for each upscaling factor, which is time-consuming and memory inefficient. We tackle this multi-scale super-resolution problem using a Filter Scaling strategy that modulates the convolution filters based on the upscaling factor, such that a single network can be used for various upscaling factors. Observing that each metabolite has distinct spatial characteristics, we also modulate the network based on the specific metabolite. Furthermore, our network is conditioned on the weight of adversarial loss so that the perceptual sharpness of the super-resolved metabolic maps can be adjusted within a single network. We incorporate these network conditionings using a novel Multi-Conditional Module. The experiments were carried out on a 1H-MRSI dataset from 15 high-grade glioma patients. Results indicate that the proposed network achieves the best performance among several multi-scale super-resolution methods and can provide super-resolved metabolic maps with adjustable sharpness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge