G. Bruce Pike
SUSEP-Net: Simulation-Supervised and Contrastive Learning-based Deep Neural Networks for Susceptibility Source Separation
Jun 16, 2025Abstract:Quantitative susceptibility mapping (QSM) provides a valuable tool for quantifying susceptibility distributions in human brains; however, two types of opposing susceptibility sources (i.e., paramagnetic and diamagnetic), may coexist in a single voxel, and cancel each other out in net QSM images. Susceptibility source separation techniques enable the extraction of sub-voxel information from QSM maps. This study proposes a novel SUSEP-Net for susceptibility source separation by training a dual-branch U-net with a simulation-supervised training strategy. In addition, a contrastive learning framework is included to explicitly impose similarity-based constraints between the branch-specific guidance features in specially-designed encoders and the latent features in the decoders. Comprehensive experiments were carried out on both simulated and in vivo data, including healthy subjects and patients with pathological conditions, to compare SUSEP-Net with three state-of-the-art susceptibility source separation methods (i.e., APART-QSM, \c{hi}-separation, and \c{hi}-sepnet). SUSEP-Net consistently showed improved results compared with the other three methods, with better numerical metrics, improved high-intensity hemorrhage and calcification lesion contrasts, and reduced artifacts in brains with pathological conditions. In addition, experiments on an agarose gel phantom data were conducted to validate the accuracy and the generalization capability of SUSEP-Net.
Highly Undersampled MRI Reconstruction via a Single Posterior Sampling of Diffusion Models
May 13, 2025Abstract:Incoherent k-space under-sampling and deep learning-based reconstruction methods have shown great success in accelerating MRI. However, the performance of most previous methods will degrade dramatically under high acceleration factors, e.g., 8$\times$ or higher. Recently, denoising diffusion models (DM) have demonstrated promising results in solving this issue; however, one major drawback of the DM methods is the long inference time due to a dramatic number of iterative reverse posterior sampling steps. In this work, a Single Step Diffusion Model-based reconstruction framework, namely SSDM-MRI, is proposed for restoring MRI images from highly undersampled k-space. The proposed method achieves one-step reconstruction by first training a conditional DM and then iteratively distilling this model. Comprehensive experiments were conducted on both publicly available fastMRI images and an in-house multi-echo GRE (QSM) subject. Overall, the results showed that SSDM-MRI outperformed other methods in terms of numerical metrics (PSNR and SSIM), qualitative error maps, image fine details, and latent susceptibility information hidden in MRI phase images. In addition, the reconstruction time for a 320*320 brain slice of SSDM-MRI is only 0.45 second, which is only comparable to that of a simple U-net, making it a highly effective solution for MRI reconstruction tasks.
Direct Estimation of Pediatric Heart Rate Variability from BOLD-fMRI: A Machine Learning Approach Using Dynamic Connectivity
Feb 10, 2025Abstract:In many pediatric fMRI studies, cardiac signals are often missing or of poor quality. A tool to extract Heart Rate Variation (HRV) waveforms directly from fMRI data, without the need for peripheral recording devices, would be highly beneficial. We developed a machine learning framework to accurately reconstruct HRV for pediatric applications. A hybrid model combining one-dimensional Convolutional Neural Networks (1D-CNN) and Gated Recurrent Units (GRU) analyzed BOLD signals from 628 ROIs, integrating past and future data. The model achieved an 8% improvement in HRV accuracy, as evidenced by enhanced performance metrics. This approach eliminates the need for peripheral photoplethysmography devices, reduces costs, and simplifies procedures in pediatric fMRI. Additionally, it improves the robustness of pediatric fMRI studies, which are more sensitive to physiological and developmental variations than those in adults.
Machine Learning-based Estimation of Respiratory Fluctuations in a Healthy Adult Population using BOLD fMRI and Head Motion Parameters
Apr 30, 2024



Abstract:Motivation: In many fMRI studies, respiratory signals are often missing or of poor quality. Therefore, it could be highly beneficial to have a tool to extract respiratory variation (RV) waveforms directly from fMRI data without the need for peripheral recording devices. Goal(s): Investigate the hypothesis that head motion parameters contain valuable information regarding respiratory patter, which can help machine learning algorithms estimate the RV waveform. Approach: This study proposes a CNN model for reconstruction of RV waveforms using head motion parameters and BOLD signals. Results: This study showed that combining head motion parameters with BOLD signals enhances RV waveform estimation. Impact: It is expected that application of the proposed method will lower the cost of fMRI studies, reduce complexity, and decrease the burden on participants as they will not be required to wear a respiratory bellows.
Plug-and-Play Latent Feature Editing for Orientation-Adaptive Quantitative Susceptibility Mapping Neural Networks
Nov 14, 2023Abstract:Quantitative susceptibility mapping (QSM) is a post-processing technique for deriving tissue magnetic susceptibility distribution from MRI phase measurements. Deep learning (DL) algorithms hold great potential for solving the ill-posed QSM reconstruction problem. However, a significant challenge facing current DL-QSM approaches is their limited adaptability to magnetic dipole field orientation variations during training and testing. In this work, we propose a novel Orientation-Adaptive Latent Feature Editing (OA-LFE) module to learn the encoding of acquisition orientation vectors and seamlessly integrate them into the latent features of deep networks. Importantly, it can be directly Plug-and-Play (PnP) into various existing DL-QSM architectures, enabling reconstructions of QSM from arbitrary magnetic dipole orientations. Its effectiveness is demonstrated by combining the OA-LFE module into our previously proposed phase-to-susceptibility single-step instant QSM (iQSM) network, which was initially tailored for pure-axial acquisitions. The proposed OA-LFE-empowered iQSM, which we refer to as iQSM+, is trained in a self-supervised manner on a specially-designed simulation brain dataset. Comprehensive experiments are conducted on simulated and in vivo human brain datasets, encompassing subjects ranging from healthy individuals to those with pathological conditions. These experiments involve various MRI platforms (3T and 7T) and aim to compare our proposed iQSM+ against several established QSM reconstruction frameworks, including the original iQSM. The iQSM+ yields QSM images with significantly improved accuracies and mitigates artifacts, surpassing other state-of-the-art DL-QSM algorithms.
Using BOLD-fMRI to Compute the Respiration Volume per Time and Respiration Variation with Convolutional Neural Networks in the Human Connectome Development Cohort
Jul 03, 2023



Abstract:In many fMRI studies, respiratory signals are unavailable or do not have acceptable quality. Consequently, the direct removal of low-frequency respiratory variations from BOLD signals is not possible. This study proposes a one-dimensional CNN model for reconstruction of two respiratory measures, RV and RVT. Results show that a CNN can capture informative features from resting BOLD signals and reconstruct realistic RV and RVT timeseries. It is expected that application of the proposed method will lower the cost of fMRI studies, reduce complexity, and decrease the burden on participants as they will not be required to wear a respiratory bellows.
Instant tissue field and magnetic susceptibility mapping from MR raw phase using Laplacian enabled deep neural networks
Nov 16, 2021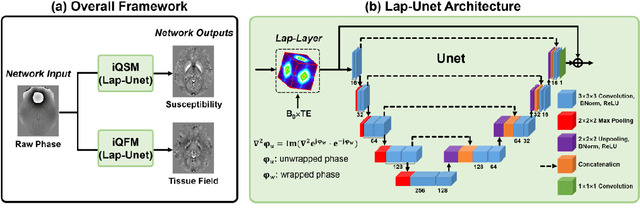
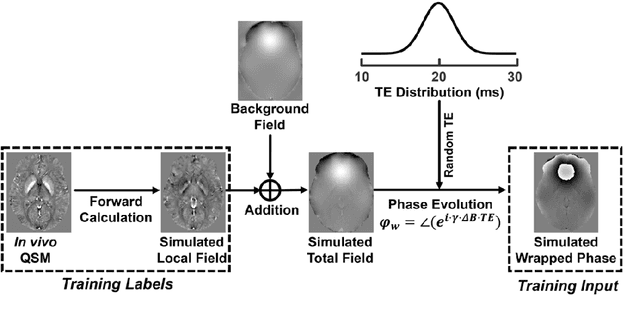


Abstract:Quantitative susceptibility mapping (QSM) is a valuable MRI post-processing technique that quantifies the magnetic susceptibility of body tissue from phase data. However, the traditional QSM reconstruction pipeline involves multiple non-trivial steps, including phase unwrapping, background field removal, and dipole inversion. These intermediate steps not only increase the reconstruction time but amplify noise and errors. This study develops a large-stencil Laplacian preprocessed deep learning-based neural network for near instant quantitative field and susceptibility mapping (i.e., iQFM and iQSM) from raw MR phase data. The proposed iQFM and iQSM methods were compared with established reconstruction pipelines on simulated and in vivo datasets. In addition, experiments on patients with intracranial hemorrhage and multiple sclerosis were also performed to test the generalization of the novel neural networks. The proposed iQFM and iQSM methods yielded comparable results to multi-step methods in healthy subjects while dramatically improving reconstruction accuracies on intracranial hemorrhages with large susceptibilities. The reconstruction time was also substantially shortened from minutes using multi-step methods to only 30 milliseconds using the trained iQFM and iQSM neural networks.
Accelerating Quantitative Susceptibility Mapping using Compressed Sensing and Deep Neural Network
Mar 17, 2021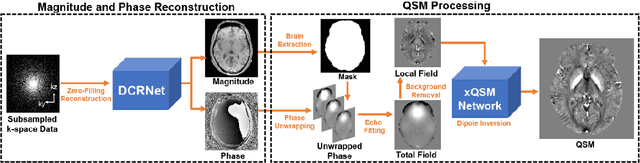
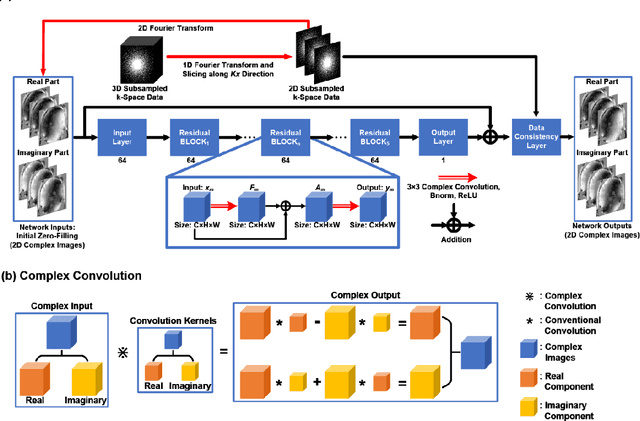
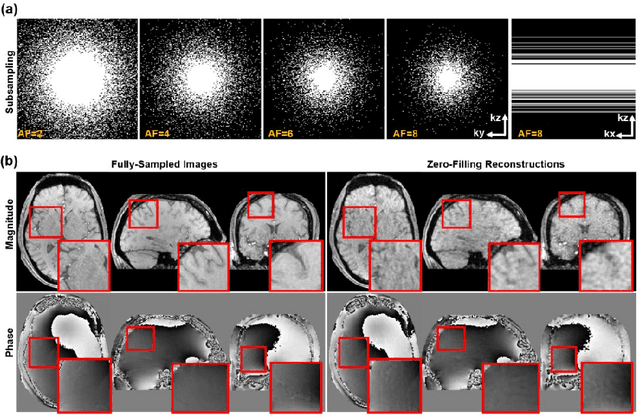
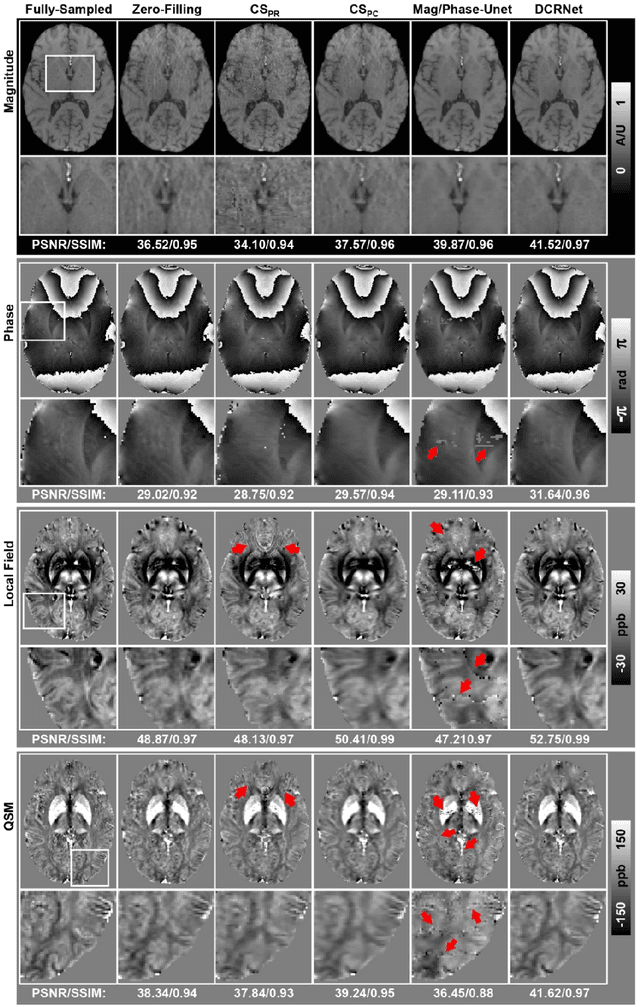
Abstract:Quantitative susceptibility mapping (QSM) is an MRI phase-based post-processing method that quantifies tissue magnetic susceptibility distributions. However, QSM acquisitions are relatively slow, even with parallel imaging. Incoherent undersampling and compressed sensing reconstruction techniques have been used to accelerate traditional magnitude-based MRI acquisitions; however, most do not recover the full phase signal due to its non-convex nature. In this study, a learning-based Deep Complex Residual Network (DCRNet) is proposed to recover both the magnitude and phase images from incoherently undersampled data, enabling high acceleration of QSM acquisition. Magnitude, phase, and QSM results from DCRNet were compared with two iterative and one deep learning methods on retrospectively undersampled acquisitions from six healthy volunteers, one intracranial hemorrhage and one multiple sclerosis patients, as well as one prospectively undersampled healthy subject using a 7T scanner. Peak signal to noise ratio (PSNR), structural similarity (SSIM) and region-of-interest susceptibility measurements are reported for numerical comparisons. The proposed DCRNet method substantially reduced artifacts and blurring compared to the other methods and resulted in the highest PSNR and SSIM on the magnitude, phase, local field, and susceptibility maps. It led to 4.0% to 8.8% accuracy improvements in deep grey matter susceptibility than some existing methods, when the acquisition was accelerated four times. The proposed DCRNet also dramatically shortened the reconstruction time by nearly 10 thousand times for each scan, from around 80 hours using conventional approaches to only 30 seconds.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge