Elias Nehme
Hierarchical Uncertainty Exploration via Feedforward Posterior Trees
May 24, 2024Abstract:When solving ill-posed inverse problems, one often desires to explore the space of potential solutions rather than be presented with a single plausible reconstruction. Valuable insights into these feasible solutions and their associated probabilities are embedded in the posterior distribution. However, when confronted with data of high dimensionality (such as images), visualizing this distribution becomes a formidable challenge, necessitating the application of effective summarization techniques before user examination. In this work, we introduce a new approach for visualizing posteriors across multiple levels of granularity using tree-valued predictions. Our method predicts a tree-valued hierarchical summarization of the posterior distribution for any input measurement, in a single forward pass of a neural network. We showcase the efficacy of our approach across diverse datasets and image restoration challenges, highlighting its prowess in uncertainty quantification and visualization. Our findings reveal that our method performs comparably to a baseline that hierarchically clusters samples from a diffusion-based posterior sampler, yet achieves this with orders of magnitude greater speed.
Uncertainty Visualization via Low-Dimensional Posterior Projections
Dec 12, 2023



Abstract:In ill-posed inverse problems, it is commonly desirable to obtain insight into the full spectrum of plausible solutions, rather than extracting only a single reconstruction. Information about the plausible solutions and their likelihoods is encoded in the posterior distribution. However, for high-dimensional data, this distribution is challenging to visualize. In this work, we introduce a new approach for estimating and visualizing posteriors by employing energy-based models (EBMs) over low-dimensional subspaces. Specifically, we train a conditional EBM that receives an input measurement and a set of directions that span some low-dimensional subspace of solutions, and outputs the probability density function of the posterior within that space. We demonstrate the effectiveness of our method across a diverse range of datasets and image restoration problems, showcasing its strength in uncertainty quantification and visualization. As we show, our method outperforms a baseline that projects samples from a diffusion-based posterior sampler, while being orders of magnitude faster. Furthermore, it is more accurate than a baseline that assumes a Gaussian posterior.
Uncertainty Quantification via Neural Posterior Principal Components
Sep 27, 2023Abstract:Uncertainty quantification is crucial for the deployment of image restoration models in safety-critical domains, like autonomous driving and biological imaging. To date, methods for uncertainty visualization have mainly focused on per-pixel estimates. However, a heatmap of per-pixel variances is typically of little practical use, as it does not capture the strong correlations between pixels. A more natural measure of uncertainty corresponds to the variances along the principal components (PCs) of the posterior distribution. Theoretically, the PCs can be computed by applying PCA on samples generated from a conditional generative model for the input image. However, this requires generating a very large number of samples at test time, which is painfully slow with the current state-of-the-art (diffusion) models. In this work, we present a method for predicting the PCs of the posterior distribution for any input image, in a single forward pass of a neural network. Our method can either wrap around a pre-trained model that was trained to minimize the mean square error (MSE), or can be trained from scratch to output both a predicted image and the posterior PCs. We showcase our method on multiple inverse problems in imaging, including denoising, inpainting, super-resolution, and biological image-to-image translation. Our method reliably conveys instance-adaptive uncertainty directions, achieving uncertainty quantification comparable with posterior samplers while being orders of magnitude faster. Examples are available at https://eliasnehme.github.io/NPPC/
Roadmap on Deep Learning for Microscopy
Mar 07, 2023



Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Learning an optimal PSF-pair for ultra-dense 3D localization microscopy
Sep 29, 2020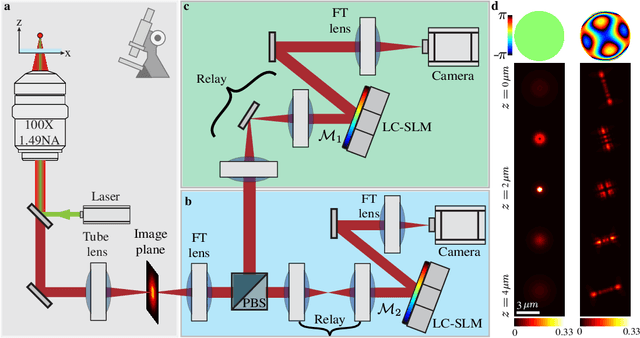
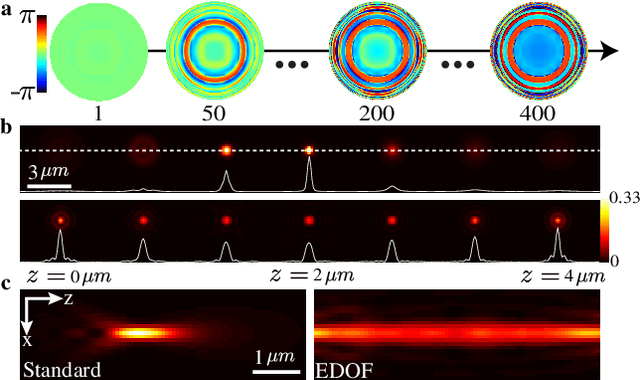
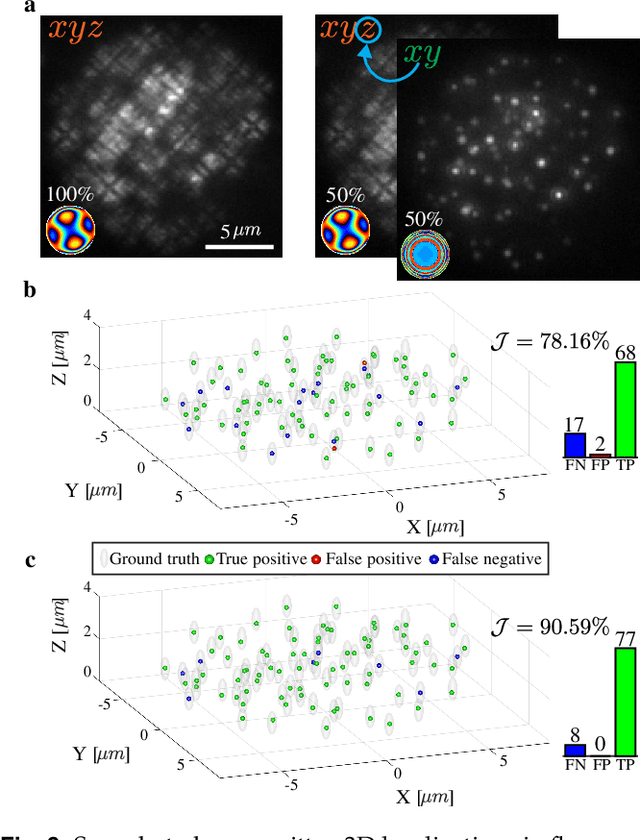
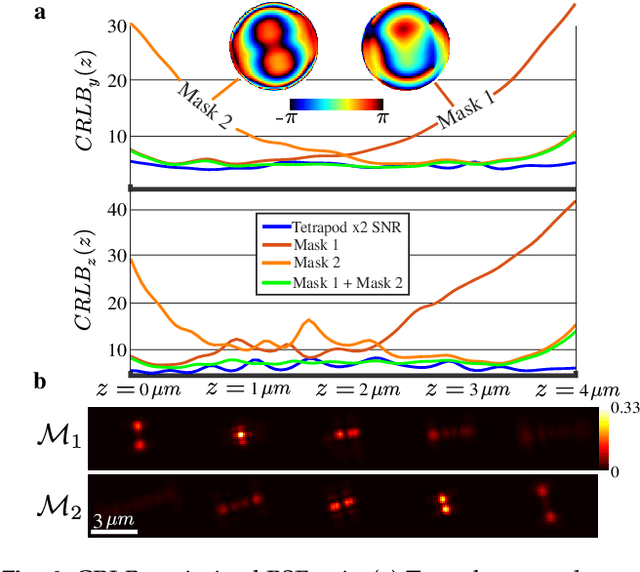
Abstract:A long-standing challenge in multiple-particle-tracking is the accurate and precise 3D localization of individual particles at close proximity. One established approach for snapshot 3D imaging is point-spread-function (PSF) engineering, in which the PSF is modified to encode the axial information. However, engineered PSFs are challenging to localize at high densities due to lateral PSF overlaps. Here we suggest using multiple PSFs simultaneously to help overcome this challenge, and investigate the problem of engineering multiple PSFs for dense 3D localization. We implement our approach using a bifurcated optical system that modifies two separate PSFs, and design the PSFs using three different approaches including end-to-end learning. We demonstrate our approach experimentally by volumetric imaging of fluorescently labelled telomeres in cells.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge