Dingkun Liu
EEG Foundation Models: Progresses, Benchmarking, and Open Problems
Jan 25, 2026Abstract:Electroencephalography (EEG) foundation models have recently emerged as a promising paradigm for brain-computer interfaces (BCIs), aiming to learn transferable neural representations from large-scale heterogeneous recordings. Despite rapid progresses, there lacks fair and comprehensive comparisons of existing EEG foundation models, due to inconsistent pre-training objectives, preprocessing choices, and downstream evaluation protocols. This paper fills this gap. We first review 50 representative models and organize their design choices into a unified taxonomic framework including data standardization, model architectures, and self-supervised pre-training strategies. We then evaluate 12 open-source foundation models and competitive specialist baselines across 13 EEG datasets spanning nine BCI paradigms. Emphasizing real-world deployments, we consider both cross-subject generalization under a leave-one-subject-out protocol and rapid calibration under a within-subject few-shot setting. We further compare full-parameter fine-tuning with linear probing to assess the transferability of pre-trained representations, and examine the relationship between model scale and downstream performance. Our results indicate that: 1) linear probing is frequently insufficient; 2) specialist models trained from scratch remain competitive across many tasks; and, 3) larger foundation models do not necessarily yield better generalization performance under current data regimes and training practices.
MIRepNet: A Pipeline and Foundation Model for EEG-Based Motor Imagery Classification
Jul 27, 2025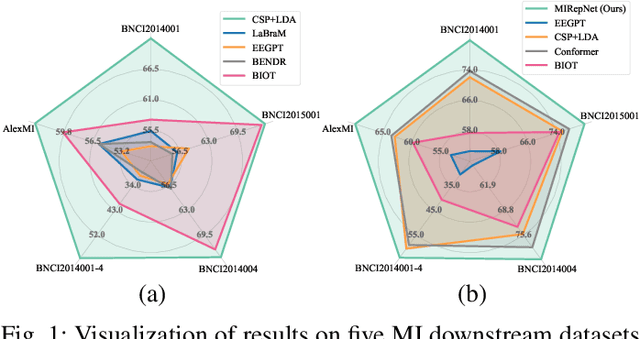



Abstract:Brain-computer interfaces (BCIs) enable direct communication between the brain and external devices. Recent EEG foundation models aim to learn generalized representations across diverse BCI paradigms. However, these approaches overlook fundamental paradigm-specific neurophysiological distinctions, limiting their generalization ability. Importantly, in practical BCI deployments, the specific paradigm such as motor imagery (MI) for stroke rehabilitation or assistive robotics, is generally determined prior to data acquisition. This paper proposes MIRepNet, the first EEG foundation model tailored for the MI paradigm. MIRepNet comprises a high-quality EEG preprocessing pipeline incorporating a neurophysiologically-informed channel template, adaptable to EEG headsets with arbitrary electrode configurations. Furthermore, we introduce a hybrid pretraining strategy that combines self-supervised masked token reconstruction and supervised MI classification, facilitating rapid adaptation and accurate decoding on novel downstream MI tasks with fewer than 30 trials per class. Extensive evaluations across five public MI datasets demonstrated that MIRepNet consistently achieved state-of-the-art performance, significantly outperforming both specialized and generalized EEG models. Our code will be available on GitHub\footnote{https://github.com/staraink/MIRepNet}.
CLEAN-MI: A Scalable and Efficient Pipeline for Constructing High-Quality Neurodata in Motor Imagery Paradigm
Jun 13, 2025Abstract:The construction of large-scale, high-quality datasets is a fundamental prerequisite for developing robust and generalizable foundation models in motor imagery (MI)-based brain-computer interfaces (BCIs). However, EEG signals collected from different subjects and devices are often plagued by low signal-to-noise ratio, heterogeneity in electrode configurations, and substantial inter-subject variability, posing significant challenges for effective model training. In this paper, we propose CLEAN-MI, a scalable and systematic data construction pipeline for constructing large-scale, efficient, and accurate neurodata in the MI paradigm. CLEAN-MI integrates frequency band filtering, channel template selection, subject screening, and marginal distribution alignment to systematically filter out irrelevant or low-quality data and standardize multi-source EEG datasets. We demonstrate the effectiveness of CLEAN-MI on multiple public MI datasets, achieving consistent improvements in data quality and classification performance.
Spatial Distillation based Distribution Alignment (SDDA) for Cross-Headset EEG Classification
Mar 07, 2025Abstract:A non-invasive brain-computer interface (BCI) enables direct interaction between the user and external devices, typically via electroencephalogram (EEG) signals. However, decoding EEG signals across different headsets remains a significant challenge due to differences in the number and locations of the electrodes. To address this challenge, we propose a spatial distillation based distribution alignment (SDDA) approach for heterogeneous cross-headset transfer in non-invasive BCIs. SDDA uses first spatial distillation to make use of the full set of electrodes, and then input/feature/output space distribution alignments to cope with the significant differences between the source and target domains. To our knowledge, this is the first work to use knowledge distillation in cross-headset transfers. Extensive experiments on six EEG datasets from two BCI paradigms demonstrated that SDDA achieved superior performance in both offline unsupervised domain adaptation and online supervised domain adaptation scenarios, consistently outperforming 10 classical and state-of-the-art transfer learning algorithms.
UMMAN: Unsupervised Multi-graph Merge Adversarial Network for Disease Prediction Based on Intestinal Flora
Jul 31, 2024



Abstract:The abundance of intestinal flora is closely related to human diseases, but diseases are not caused by a single gut microbe. Instead, they result from the complex interplay of numerous microbial entities. This intricate and implicit connection among gut microbes poses a significant challenge for disease prediction using abundance information from OTU data. Recently, several methods have shown potential in predicting corresponding diseases. However, these methods fail to learn the inner association among gut microbes from different hosts, leading to unsatisfactory performance. In this paper, we present a novel architecture, Unsupervised Multi-graph Merge Adversarial Network (UMMAN). UMMAN can obtain the embeddings of nodes in the Multi-Graph in an unsupervised scenario, so that it helps learn the multiplex association. Our method is the first to combine Graph Neural Network with the task of intestinal flora disease prediction. We employ complex relation-types to construct the Original-Graph and disrupt the relationships among nodes to generate corresponding Shuffled-Graph. We introduce the Node Feature Global Integration (NFGI) module to represent the global features of the graph. Furthermore, we design a joint loss comprising adversarial loss and hybrid attention loss to ensure that the real graph embedding aligns closely with the Original-Graph and diverges from the Shuffled-Graph. Comprehensive experiments on five classical OTU gut microbiome datasets demonstrate the effectiveness and stability of our method. (We will release our code soon.)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge