David Rolnick
Mila Quebec AI Institute, Canada, McGill University, Canada
Application-Driven Innovation in Machine Learning
Mar 26, 2024


Abstract:As applications of machine learning proliferate, innovative algorithms inspired by specific real-world challenges have become increasingly important. Such work offers the potential for significant impact not merely in domains of application but also in machine learning itself. In this paper, we describe the paradigm of application-driven research in machine learning, contrasting it with the more standard paradigm of methods-driven research. We illustrate the benefits of application-driven machine learning and how this approach can productively synergize with methods-driven work. Despite these benefits, we find that reviewing, hiring, and teaching practices in machine learning often hold back application-driven innovation. We outline how these processes may be improved.
Dataset Difficulty and the Role of Inductive Bias
Jan 03, 2024Abstract:Motivated by the goals of dataset pruning and defect identification, a growing body of methods have been developed to score individual examples within a dataset. These methods, which we call "example difficulty scores", are typically used to rank or categorize examples, but the consistency of rankings between different training runs, scoring methods, and model architectures is generally unknown. To determine how example rankings vary due to these random and controlled effects, we systematically compare different formulations of scores over a range of runs and model architectures. We find that scores largely share the following traits: they are noisy over individual runs of a model, strongly correlated with a single notion of difficulty, and reveal examples that range from being highly sensitive to insensitive to the inductive biases of certain model architectures. Drawing from statistical genetics, we develop a simple method for fingerprinting model architectures using a few sensitive examples. These findings guide practitioners in maximizing the consistency of their scores (e.g. by choosing appropriate scoring methods, number of runs, and subsets of examples), and establishes comprehensive baselines for evaluating scores in the future.
FoMo-Bench: a multi-modal, multi-scale and multi-task Forest Monitoring Benchmark for remote sensing foundation models
Dec 15, 2023



Abstract:Forests are an essential part of Earth's ecosystems and natural systems, as well as providing services on which humanity depends, yet they are rapidly changing as a result of land use decisions and climate change. Understanding and mitigating negative effects requires parsing data on forests at global scale from a broad array of sensory modalities, and recently many such problems have been approached using machine learning algorithms for remote sensing. To date, forest-monitoring problems have largely been approached in isolation. Inspired by the rise of foundation models for computer vision and remote sensing, we here present the first unified Forest Monitoring Benchmark (FoMo-Bench). FoMo-Bench consists of 15 diverse datasets encompassing satellite, aerial, and inventory data, covering a variety of geographical regions, and including multispectral, red-green-blue, synthetic aperture radar (SAR) and LiDAR data with various temporal, spatial and spectral resolutions. FoMo-Bench includes multiple types of forest-monitoring tasks, spanning classification, segmentation, and object detection. To further enhance the diversity of tasks and geographies represented in FoMo-Bench, we introduce a novel global dataset, TalloS, combining satellite imagery with ground-based annotations for tree species classification, spanning 1,000+ hierarchical taxonomic levels (species, genus, family). Finally, we propose FoMo-Net, a foundation model baseline designed for forest monitoring with the flexibility to process any combination of commonly used sensors in remote sensing. This work aims to inspire research collaborations between machine learning and forest biology researchers in exploring scalable multi-modal and multi-task models for forest monitoring. All code and data will be made publicly available.
Towards Causal Representations of Climate Model Data
Dec 06, 2023

Abstract:Climate models, such as Earth system models (ESMs), are crucial for simulating future climate change based on projected Shared Socioeconomic Pathways (SSP) greenhouse gas emissions scenarios. While ESMs are sophisticated and invaluable, machine learning-based emulators trained on existing simulation data can project additional climate scenarios much faster and are computationally efficient. However, they often lack generalizability and interpretability. This work delves into the potential of causal representation learning, specifically the \emph{Causal Discovery with Single-parent Decoding} (CDSD) method, which could render climate model emulation efficient \textit{and} interpretable. We evaluate CDSD on multiple climate datasets, focusing on emissions, temperature, and precipitation. Our findings shed light on the challenges, limitations, and promise of using CDSD as a stepping stone towards more interpretable and robust climate model emulation.
ClimateSet: A Large-Scale Climate Model Dataset for Machine Learning
Nov 07, 2023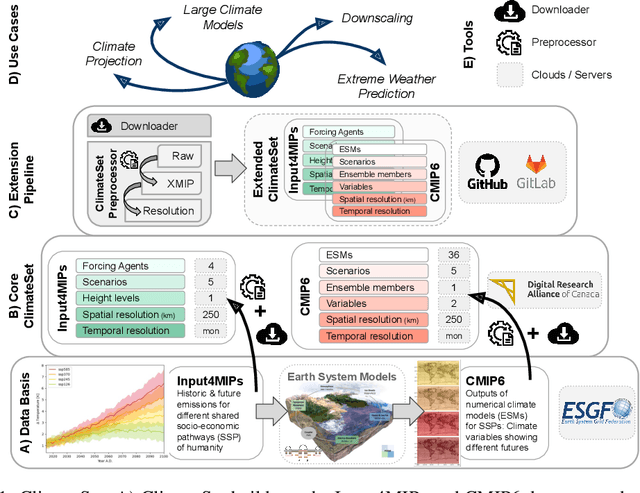
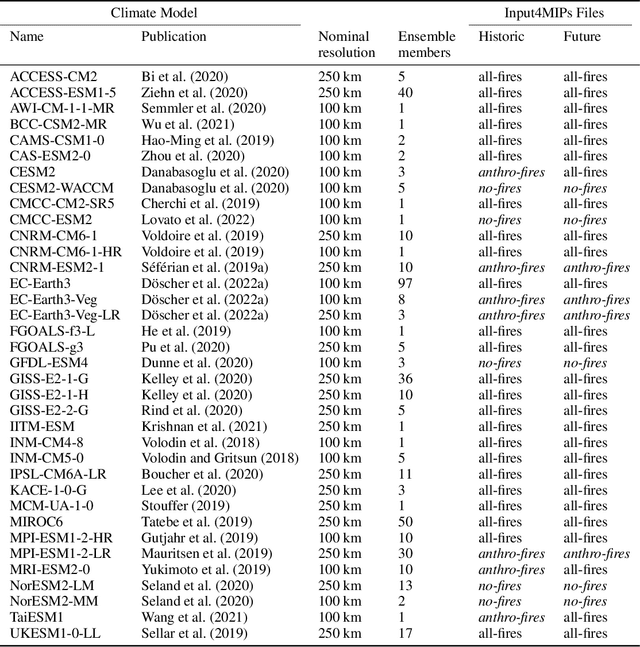
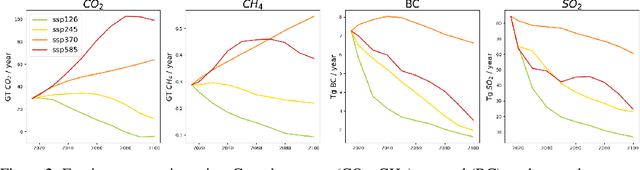
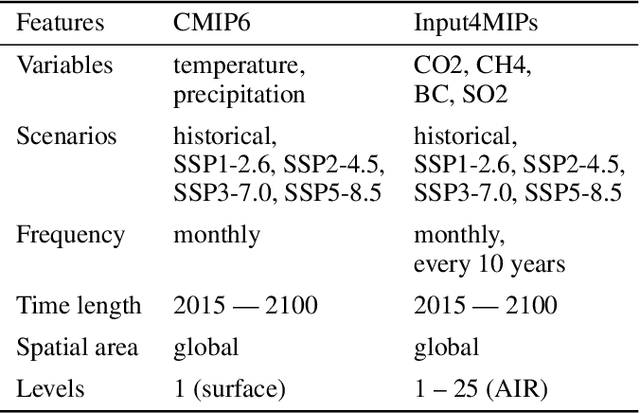
Abstract:Climate models have been key for assessing the impact of climate change and simulating future climate scenarios. The machine learning (ML) community has taken an increased interest in supporting climate scientists' efforts on various tasks such as climate model emulation, downscaling, and prediction tasks. Many of those tasks have been addressed on datasets created with single climate models. However, both the climate science and ML communities have suggested that to address those tasks at scale, we need large, consistent, and ML-ready climate model datasets. Here, we introduce ClimateSet, a dataset containing the inputs and outputs of 36 climate models from the Input4MIPs and CMIP6 archives. In addition, we provide a modular dataset pipeline for retrieving and preprocessing additional climate models and scenarios. We showcase the potential of our dataset by using it as a benchmark for ML-based climate model emulation. We gain new insights about the performance and generalization capabilities of the different ML models by analyzing their performance across different climate models. Furthermore, the dataset can be used to train an ML emulator on several climate models instead of just one. Such a "super emulator" can quickly project new climate change scenarios, complementing existing scenarios already provided to policymakers. We believe ClimateSet will create the basis needed for the ML community to tackle climate-related tasks at scale.
OpenForest: A data catalogue for machine learning in forest monitoring
Nov 02, 2023Abstract:Forests play a crucial role in Earth's system processes and provide a suite of social and economic ecosystem services, but are significantly impacted by human activities, leading to a pronounced disruption of the equilibrium within ecosystems. Advancing forest monitoring worldwide offers advantages in mitigating human impacts and enhancing our comprehension of forest composition, alongside the effects of climate change. While statistical modeling has traditionally found applications in forest biology, recent strides in machine learning and computer vision have reached important milestones using remote sensing data, such as tree species identification, tree crown segmentation and forest biomass assessments. For this, the significance of open access data remains essential in enhancing such data-driven algorithms and methodologies. Here, we provide a comprehensive and extensive overview of 86 open access forest datasets across spatial scales, encompassing inventories, ground-based, aerial-based, satellite-based recordings, and country or world maps. These datasets are grouped in OpenForest, a dynamic catalogue open to contributions that strives to reference all available open access forest datasets. Moreover, in the context of these datasets, we aim to inspire research in machine learning applied to forest biology by establishing connections between contemporary topics, perspectives and challenges inherent in both domains. We hope to encourage collaborations among scientists, fostering the sharing and exploration of diverse datasets through the application of machine learning methods for large-scale forest monitoring. OpenForest is available at https://github.com/RolnickLab/OpenForest .
SatBird: Bird Species Distribution Modeling with Remote Sensing and Citizen Science Data
Nov 02, 2023



Abstract:Biodiversity is declining at an unprecedented rate, impacting ecosystem services necessary to ensure food, water, and human health and well-being. Understanding the distribution of species and their habitats is crucial for conservation policy planning. However, traditional methods in ecology for species distribution models (SDMs) generally focus either on narrow sets of species or narrow geographical areas and there remain significant knowledge gaps about the distribution of species. A major reason for this is the limited availability of data traditionally used, due to the prohibitive amount of effort and expertise required for traditional field monitoring. The wide availability of remote sensing data and the growing adoption of citizen science tools to collect species observations data at low cost offer an opportunity for improving biodiversity monitoring and enabling the modelling of complex ecosystems. We introduce a novel task for mapping bird species to their habitats by predicting species encounter rates from satellite images, and present SatBird, a satellite dataset of locations in the USA with labels derived from presence-absence observation data from the citizen science database eBird, considering summer (breeding) and winter seasons. We also provide a dataset in Kenya representing low-data regimes. We additionally provide environmental data and species range maps for each location. We benchmark a set of baselines on our dataset, including SOTA models for remote sensing tasks. SatBird opens up possibilities for scalably modelling properties of ecosystems worldwide.
On the importance of catalyst-adsorbate 3D interactions for relaxed energy predictions
Oct 10, 2023Abstract:The use of machine learning for material property prediction and discovery has traditionally centered on graph neural networks that incorporate the geometric configuration of all atoms. However, in practice not all this information may be readily available, e.g.~when evaluating the potentially unknown binding of adsorbates to catalyst. In this paper, we investigate whether it is possible to predict a system's relaxed energy in the OC20 dataset while ignoring the relative position of the adsorbate with respect to the electro-catalyst. We consider SchNet, DimeNet++ and FAENet as base architectures and measure the impact of four modifications on model performance: removing edges in the input graph, pooling independent representations, not sharing the backbone weights and using an attention mechanism to propagate non-geometric relative information. We find that while removing binding site information impairs accuracy as expected, modified models are able to predict relaxed energies with remarkably decent MAE. Our work suggests future research directions in accelerated materials discovery where information on reactant configurations can be reduced or altogether omitted.
Multi-variable Hard Physical Constraints for Climate Model Downscaling
Aug 02, 2023


Abstract:Global Climate Models (GCMs) are the primary tool to simulate climate evolution and assess the impacts of climate change. However, they often operate at a coarse spatial resolution that limits their accuracy in reproducing local-scale phenomena. Statistical downscaling methods leveraging deep learning offer a solution to this problem by approximating local-scale climate fields from coarse variables, thus enabling regional GCM projections. Typically, climate fields of different variables of interest are downscaled independently, resulting in violations of fundamental physical properties across interconnected variables. This study investigates the scope of this problem and, through an application on temperature, lays the foundation for a framework introducing multi-variable hard constraints that guarantees physical relationships between groups of downscaled climate variables.
Hidden symmetries of ReLU networks
Jun 09, 2023Abstract:The parameter space for any fixed architecture of feedforward ReLU neural networks serves as a proxy during training for the associated class of functions - but how faithful is this representation? It is known that many different parameter settings can determine the same function. Moreover, the degree of this redundancy is inhomogeneous: for some networks, the only symmetries are permutation of neurons in a layer and positive scaling of parameters at a neuron, while other networks admit additional hidden symmetries. In this work, we prove that, for any network architecture where no layer is narrower than the input, there exist parameter settings with no hidden symmetries. We also describe a number of mechanisms through which hidden symmetries can arise, and empirically approximate the functional dimension of different network architectures at initialization. These experiments indicate that the probability that a network has no hidden symmetries decreases towards 0 as depth increases, while increasing towards 1 as width and input dimension increase.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge