David Carlson
ClinStructor: AI-Powered Structuring of Unstructured Clinical Texts
Nov 14, 2025



Abstract:Clinical notes contain valuable, context-rich information, but their unstructured format introduces several challenges, including unintended biases (e.g., gender or racial bias), and poor generalization across clinical settings (e.g., models trained on one EHR system may perform poorly on another due to format differences) and poor interpretability. To address these issues, we present ClinStructor, a pipeline that leverages large language models (LLMs) to convert clinical free-text into structured, task-specific question-answer pairs prior to predictive modeling. Our method substantially enhances transparency and controllability and only leads to a modest reduction in predictive performance (a 2-3% drop in AUC), compared to direct fine-tuning, on the ICU mortality prediction task. ClinStructor lays a strong foundation for building reliable, interpretable, and generalizable machine learning models in clinical environments.
Additive Large Language Models for Semi-Structured Text
Nov 14, 2025Abstract:Large Language Models have advanced clinical text classification, but their opaque predictions remain a critical barrier to practical adoption in research and clinical settings where investigators and physicians need to understand which parts of a patient's record drive risk signals. To address this challenge, we introduce \textbf{CALM}, short for \textbf{Classification with Additive Large Language Models}, an interpretable framework for semi-structured text where inputs are composed of semantically meaningful components, such as sections of an admission note or question-answer fields from an intake form. CALM predicts outcomes as the additive sum of each component's contribution, making these contributions part of the forward computation itself and enabling faithful explanations at both the patient and population level. The additive structure also enables clear visualizations, such as component-level risk curves similar to those used in generalized additive models, making the learned relationships easier to inspect and communicate. Although CALM expects semi-structured inputs, many clinical documents already have this form, and similar structure can often be automatically extracted from free-text notes. CALM achieves performance comparable to conventional LLM classifiers while improving trust, supporting quality-assurance checks, and revealing clinically meaningful patterns during model development and auditing.
Generating Hypotheses of Dynamic Causal Graphs in Neuroscience: Leveraging Generative Factor Models of Observed Time Series
May 27, 2025Abstract:The field of hypothesis generation promises to reduce costs in neuroscience by narrowing the range of interventional studies needed to study various phenomena. Existing machine learning methods can generate scientific hypotheses from complex datasets, but many approaches assume causal relationships are static over time, limiting their applicability to systems with dynamic, state-dependent behavior, such as the brain. While some techniques attempt dynamic causal discovery through factor models, they often restrict relationships to linear patterns or impose other simplifying assumptions. We propose a novel method that models dynamic graphs as a conditionally weighted superposition of static graphs, where each static graph can capture nonlinear relationships. This approach enables the detection of complex, time-varying interactions between variables beyond linear limitations. Our method improves f1-scores of predicted dynamic causal patterns by roughly 22-28% on average over baselines in some of our experiments, with some improvements reaching well over 60%. A case study on real brain data demonstrates our method's ability to uncover relationships linked to specific behavioral states, offering valuable insights into neural dynamics.
Deep Causal Inference for Point-referenced Spatial Data with Continuous Treatments
Dec 05, 2024



Abstract:Causal reasoning is often challenging with spatial data, particularly when handling high-dimensional inputs. To address this, we propose a neural network (NN) based framework integrated with an approximate Gaussian process to manage spatial interference and unobserved confounding. Additionally, we adopt a generalized propensity-score-based approach to address partially observed outcomes when estimating causal effects with continuous treatments. We evaluate our framework using synthetic, semi-synthetic, and real-world data inferred from satellite imagery. Our results demonstrate that NN-based models significantly outperform linear spatial regression models in estimating causal effects. Furthermore, in real-world case studies, NN-based models offer more reasonable predictions of causal effects, facilitating decision-making in relevant applications.
Augmenting Ground-Level PM2.5 Prediction via Kriging-Based Pseudo-Label Generation
Jan 16, 2024Abstract:Fusing abundant satellite data with sparse ground measurements constitutes a major challenge in climate modeling. To address this, we propose a strategy to augment the training dataset by introducing unlabeled satellite images paired with pseudo-labels generated through a spatial interpolation technique known as ordinary kriging, thereby making full use of the available satellite data resources. We show that the proposed data augmentation strategy helps enhance the performance of the state-of-the-art convolutional neural network-random forest (CNN-RF) model by a reasonable amount, resulting in a noteworthy improvement in spatial correlation and a reduction in prediction error.
Causal Mediation Analysis with Multi-dimensional and Indirectly Observed Mediators
Jun 13, 2023



Abstract:Causal mediation analysis (CMA) is a powerful method to dissect the total effect of a treatment into direct and mediated effects within the potential outcome framework. This is important in many scientific applications to identify the underlying mechanisms of a treatment effect. However, in many scientific applications the mediator is unobserved, but there may exist related measurements. For example, we may want to identify how changes in brain activity or structure mediate an antidepressant's effect on behavior, but we may only have access to electrophysiological or imaging brain measurements. To date, most CMA methods assume that the mediator is one-dimensional and observable, which oversimplifies such real-world scenarios. To overcome this limitation, we introduce a CMA framework that can handle complex and indirectly observed mediators based on the identifiable variational autoencoder (iVAE) architecture. We prove that the true joint distribution over observed and latent variables is identifiable with the proposed method. Additionally, our framework captures a disentangled representation of the indirectly observed mediator and yields accurate estimation of the direct and mediated effects in synthetic and semi-synthetic experiments, providing evidence of its potential utility in real-world applications.
Domain Adaptation via Rebalanced Sub-domain Alignment
Feb 03, 2023



Abstract:Unsupervised domain adaptation (UDA) is a technique used to transfer knowledge from a labeled source domain to a different but related unlabeled target domain. While many UDA methods have shown success in the past, they often assume that the source and target domains must have identical class label distributions, which can limit their effectiveness in real-world scenarios. To address this limitation, we propose a novel generalization bound that reweights source classification error by aligning source and target sub-domains. We prove that our proposed generalization bound is at least as strong as existing bounds under realistic assumptions, and we empirically show that it is much stronger on real-world data. We then propose an algorithm to minimize this novel generalization bound. We demonstrate by numerical experiments that this approach improves performance in shifted class distribution scenarios compared to state-of-the-art methods.
Estimating Causal Effects using a Multi-task Deep Ensemble
Jan 26, 2023



Abstract:Over the past few decades, a number of methods have been proposed for causal effect estimation, yet few have been demonstrated to be effective in handling data with complex structures, such as images. To fill this gap, we propose a Causal Multi-task Deep Ensemble (CMDE) framework to learn both shared and group-specific information from the study population and prove its equivalence to a multi-task Gaussian process (GP) with coregionalization kernel a priori. Compared to multi-task GP, CMDE efficiently handles high-dimensional and multi-modal covariates and provides pointwise uncertainty estimates of causal effects. We evaluate our method across various types of datasets and tasks and find that CMDE outperforms state-of-the-art methods on a majority of these tasks.
Incorporating Prior Knowledge into Neural Networks through an Implicit Composite Kernel
May 17, 2022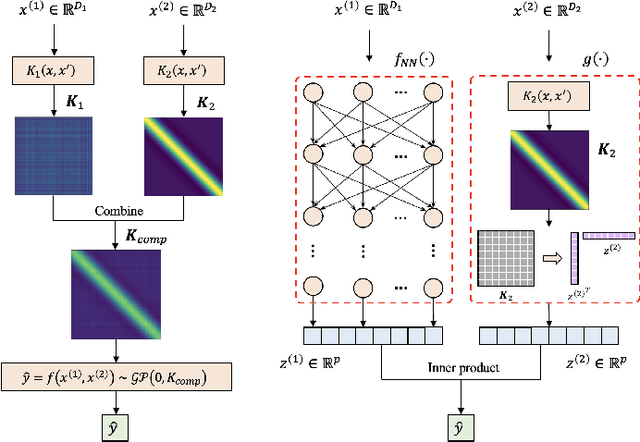

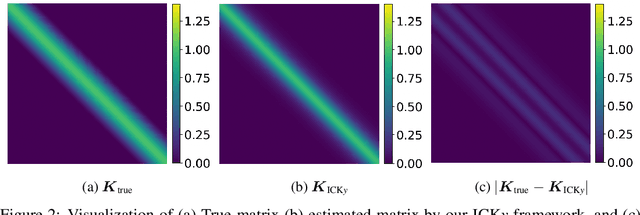
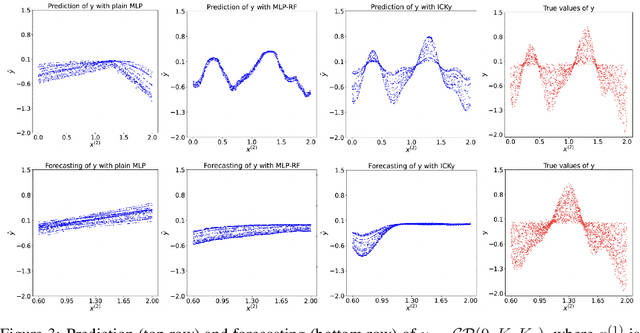
Abstract:It is challenging to guide neural network (NN) learning with prior knowledge. In contrast, many known properties, such as spatial smoothness or seasonality, are straightforward to model by choosing an appropriate kernel in a Gaussian process (GP). Many deep learning applications could be enhanced by modeling such known properties. For example, convolutional neural networks (CNNs) are frequently used in remote sensing, which is subject to strong seasonal effects. We propose to blend the strengths of deep learning and the clear modeling capabilities of GPs by using a composite kernel that combines a kernel implicitly defined by a neural network with a second kernel function chosen to model known properties (e.g., seasonality). Then, we approximate the resultant GP by combining a deep network and an efficient mapping based on the Nystrom approximation, which we call Implicit Composite Kernel (ICK). ICK is flexible and can be used to include prior information in neural networks in many applications. We demonstrate the strength of our framework by showing its superior performance and flexibility on both synthetic and real-world data sets. The code is available at: https://anonymous.4open.science/r/ICK_NNGP-17C5/.
Multiple Domain Causal Networks
May 13, 2022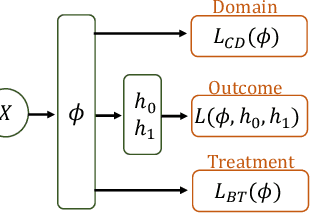
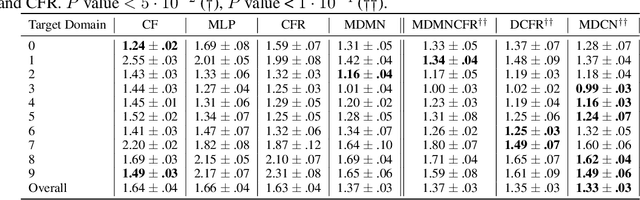
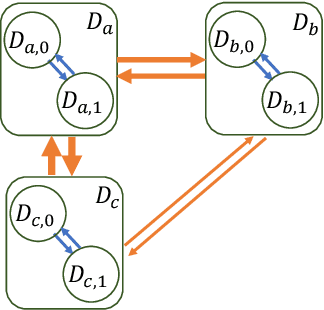
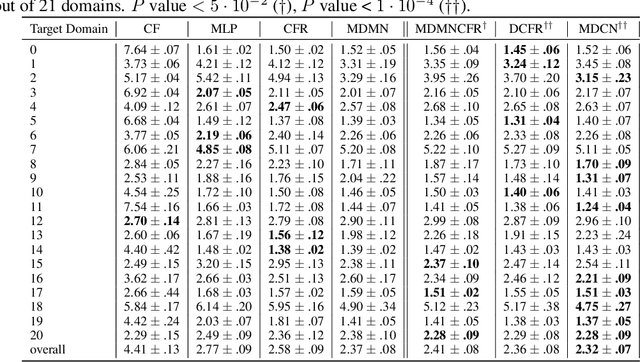
Abstract:Observational studies are regarded as economic alternatives to randomized trials, often used in their stead to investigate and determine treatment efficacy. Due to lack of sample size, observational studies commonly combine data from multiple sources or different sites/centers. Despite the benefits of an increased sample size, a naive combination of multicenter data may result in incongruities stemming from center-specific protocols for generating cohorts or reactions towards treatments distinct to a given center, among other things. These issues arise in a variety of other contexts, including capturing a treatment effect related to an individual's unique biological characteristics. Existing methods for estimating heterogeneous treatment effects have not adequately addressed the multicenter context, but rather treat it simply as a means to obtain sufficient sample size. Additionally, previous approaches to estimating treatment effects do not straightforwardly generalize to the multicenter design, especially when required to provide treatment insights for patients from a new, unobserved center. To address these shortcomings, we propose Multiple Domain Causal Networks (MDCN), an approach that simultaneously strengthens the information sharing between similar centers while addressing the selection bias in treatment assignment through learning of a new feature embedding. In empirical evaluations, MDCN is consistently more accurate when estimating the heterogeneous treatment effect in new centers compared to benchmarks that adjust solely based on treatment imbalance or general center differences. Finally, we justify our approach by providing theoretical analyses that demonstrate that MDCN improves on the generalization bound of the new, unobserved target center.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge