Dario Augusto Borges Oliveira
Evolution of urban areas and land surface temperature
Jan 05, 2024Abstract:With the global population on the rise, our cities have been expanding to accommodate the growing number of people. The expansion of cities generally leads to the engulfment of peripheral areas. However, such expansion of urban areas is likely to cause increment in areas with increased land surface temperature (LST). By considering each summer as a data point, we form LST multi-year time-series and cluster it to obtain spatio-temporal pattern. We observe several interesting phenomena from these patterns, e.g., some clusters show reasonable similarity to the built-up area, whereas the locations with high temporal variation are seen more in the peripheral areas. Furthermore, the LST center of mass shifts over the years for cities with development activities tilted towards a direction. We conduct the above-mentioned studies for three different cities in three different continents.
Exploring Self-Attention for Crop-type Classification Explainability
Oct 24, 2022



Abstract:Automated crop-type classification using Sentinel-2 satellite time series is essential to support agriculture monitoring. Recently, deep learning models based on transformer encoders became a promising approach for crop-type classification. Using explainable machine learning to reveal the inner workings of these models is an important step towards improving stakeholders' trust and efficient agriculture monitoring. In this paper, we introduce a novel explainability framework that aims to shed a light on the essential crop disambiguation patterns learned by a state-of-the-art transformer encoder model. More specifically, we process the attention weights of a trained transformer encoder to reveal the critical dates for crop disambiguation and use domain knowledge to uncover the phenological events that support the model performance. We also present a sensitivity analysis approach to understand better the attention capability for revealing crop-specific phenological events. We report compelling results showing that attention patterns strongly relate to key dates, and consequently, to the critical phenological events for crop-type classification. These findings might be relevant for improving stakeholder trust and optimizing agriculture monitoring processes. Additionally, our sensitivity analysis demonstrates the limitation of attention weights for identifying the important events in the crop phenology as we empirically show that the unveiled phenological events depend on the other crops in the data considered during training.
Controlling Weather Field Synthesis Using Variational Autoencoders
Jul 30, 2021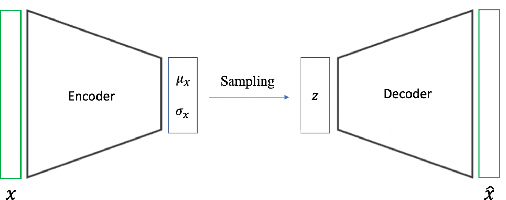
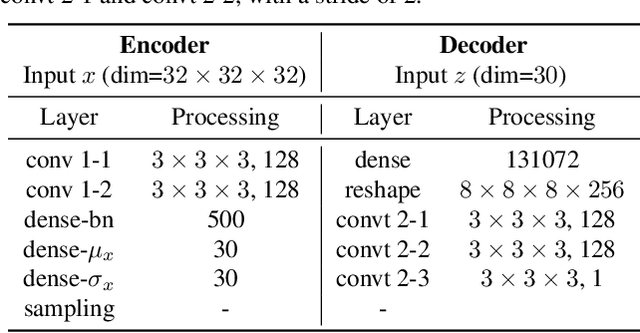
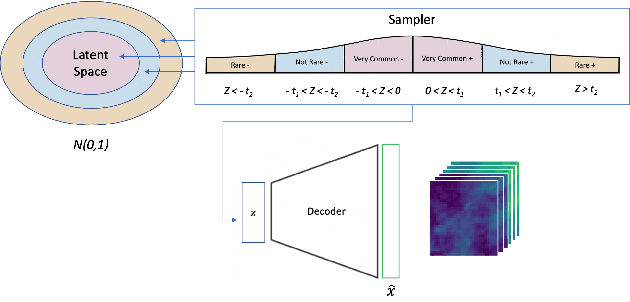
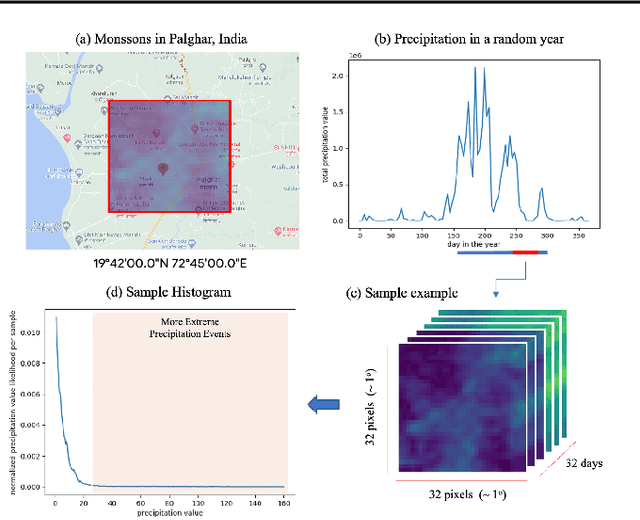
Abstract:One of the consequences of climate change is anobserved increase in the frequency of extreme cli-mate events. That poses a challenge for weatherforecast and generation algorithms, which learnfrom historical data but should embed an often un-certain bias to create correct scenarios. This paperinvestigates how mapping climate data to a knowndistribution using variational autoencoders mighthelp explore such biases and control the synthesisof weather fields towards more extreme climatescenarios. We experimented using a monsoon-affected precipitation dataset from southwest In-dia, which should give a roughly stable pattern ofrainy days and ease our investigation. We reportcompelling results showing that mapping complexweather data to a known distribution implementsan efficient control for weather field synthesis to-wards more (or less) extreme scenarios.
Multi-task fully convolutional network for tree species mapping in dense forests using small training hyperspectral data
Jun 01, 2021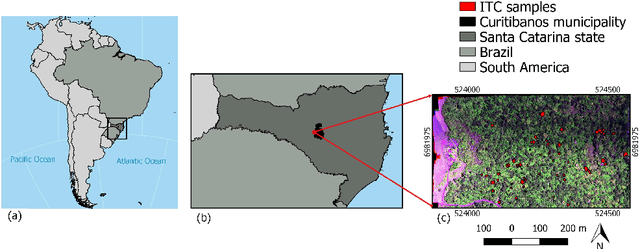
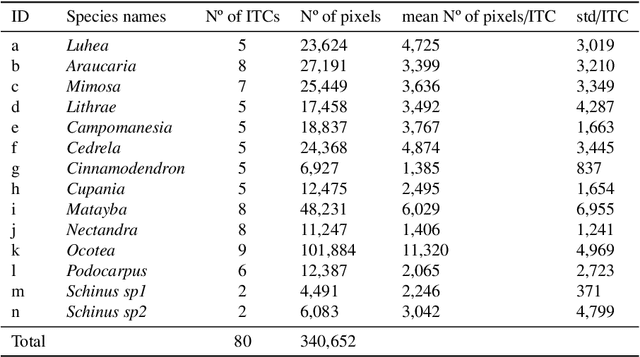

Abstract:This work proposes a multi-task fully convolutional architecture for tree species mapping in dense forests from sparse and scarce polygon-level annotations using hyperspectral UAV-borne data. Our model implements a partial loss function that enables dense tree semantic labeling outcomes from non-dense training samples, and a distance regression complementary task that enforces tree crown boundary constraints and substantially improves the model performance. Our multi-task architecture uses a shared backbone network that learns common representations for both tasks and two task-specific decoders, one for the semantic segmentation output and one for the distance map regression. We report that introducing the complementary task boosts the semantic segmentation performance compared to the single-task counterpart in up to 10% reaching an overall F1 score of 87.5% and an overall accuracy of 85.9%, achieving state-of-art performance for tree species classification in tropical forests.
Decoupling Shape and Density for Liver Lesion Synthesis Using Conditional Generative Adversarial Networks
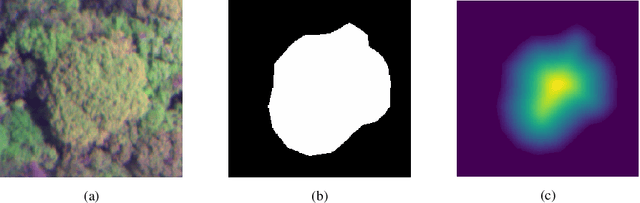
Jun 01, 2021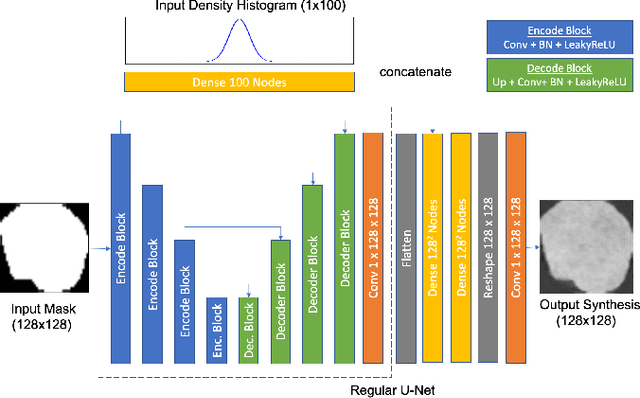

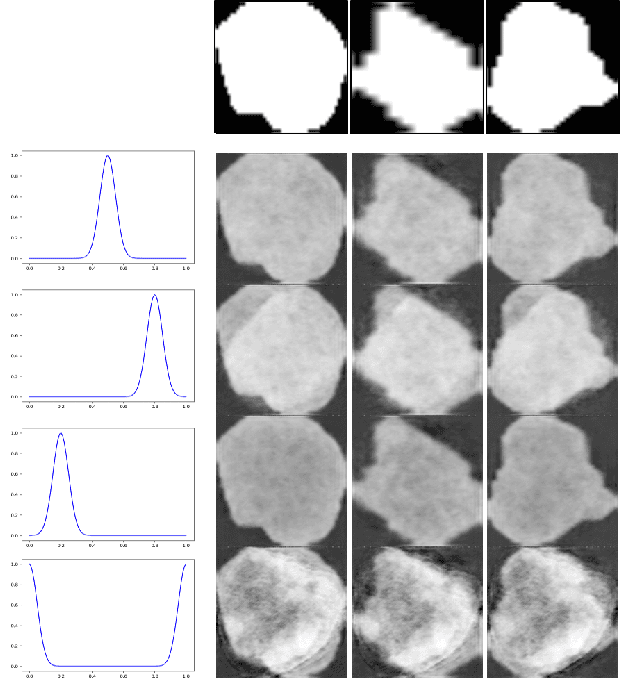
Abstract:Lesion synthesis received much attention with the rise of efficient generative models for augmenting training data, drawing lesion evolution scenarios, or aiding expert training. The quality and diversity of synthesized data are highly dependent on the annotated data used to train the models, which not rarely struggle to derive very different yet realistic samples from the training ones. That adds an inherent bias to lesion segmentation algorithms and limits synthesizing lesion evolution scenarios efficiently. This paper presents a method for decoupling shape and density for liver lesion synthesis, creating a framework that allows straight-forwardly driving the synthesis. We offer qualitative results that show the synthesis control by modifying shape and density individually, and quantitative results that demonstrate that embedding the density information in the generator model helps to increase lesion segmentation performance compared to using the shape solely.
Ground Roll Suppression using Convolutional Neural Networks
Oct 28, 2020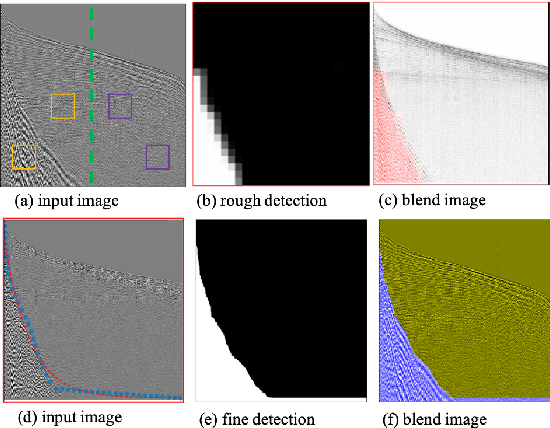
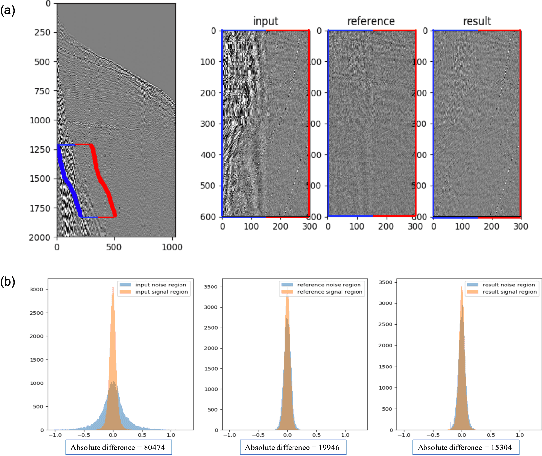
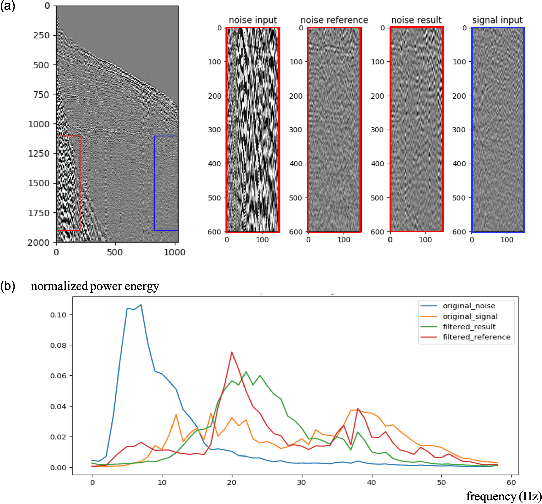
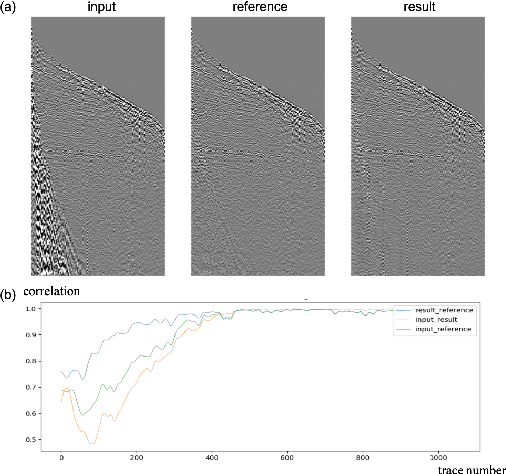
Abstract:Seismic data processing plays a major role in seismic exploration as it conditions much of the seismic interpretation performance. In this context, generating reliable post-stack seismic data depends also on disposing of an efficient pre-stack noise attenuation tool. Here we tackle ground roll noise, one of the most challenging and common noises observed in pre-stack seismic data. Since ground roll is characterized by relative low frequencies and high amplitudes, most commonly used approaches for its suppression are based on frequency-amplitude filters for ground roll characteristic bands. However, when signal and noise share the same frequency ranges, these methods usually deliver also signal suppression or residual noise. In this paper we take advantage of the highly non-linear features of convolutional neural networks, and propose to use different architectures to detect ground roll in shot gathers and ultimately to suppress them using conditional generative adversarial networks. Additionally, we propose metrics to evaluate ground roll suppression, and report strong results compared to expert filtering. Finally, we discuss generalization of trained models for similar and different geologies to better understand the feasibility of our proposal in real applications.
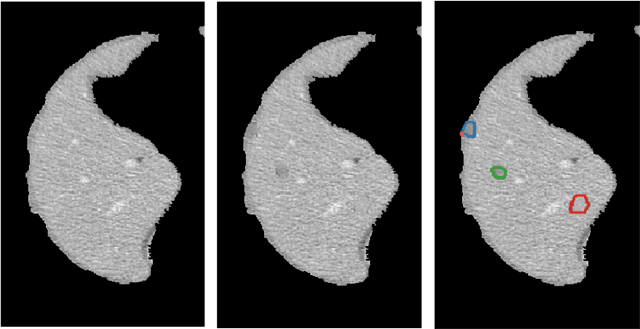
Implanting Synthetic Lesions for Improving Liver Lesion Segmentation in CT Exams
Aug 11, 2020
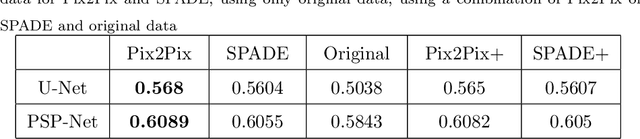
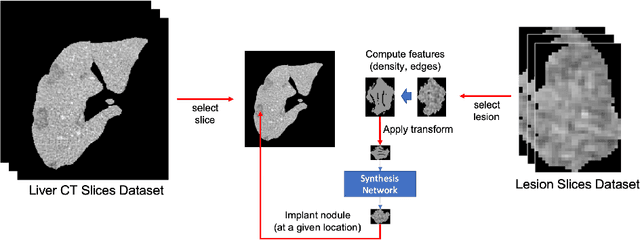
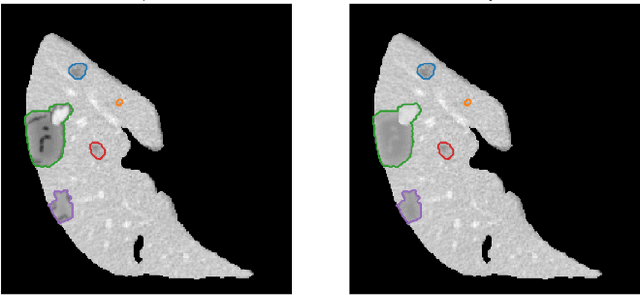
Abstract:The success of supervised lesion segmentation algorithms using Computed Tomography (CT) exams depends significantly on the quantity and variability of samples available for training. While annotating such data constitutes a challenge itself, the variability of lesions in the dataset also depends on the prevalence of different types of lesions. This phenomenon adds an inherent bias to lesion segmentation algorithms that can be diminished, among different possibilities, using aggressive data augmentation methods. In this paper, we present a method for implanting realistic lesions in CT slices to provide a rich and controllable set of training samples and ultimately improving semantic segmentation network performances for delineating lesions in CT exams. Our results show that implanting synthetic lesions not only improves (up to around 12\%) the segmentation performance considering different architectures but also that this improvement is consistent among different image synthesis networks. We conclude that increasing the variability of lesions synthetically in terms of size, density, shape, and position seems to improve the performance of segmentation models for liver lesion segmentation in CT slices.
An argument in favor of strong scaling for deep neural networks with small datasets
Sep 25, 2018
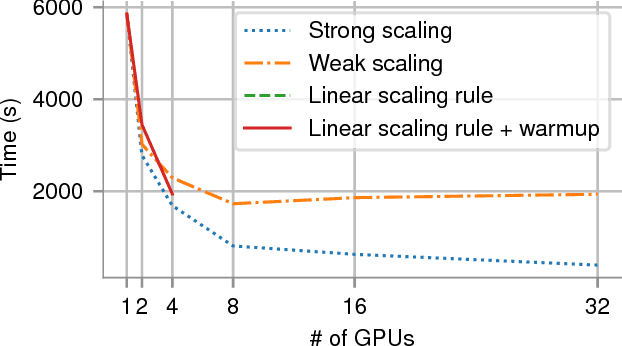
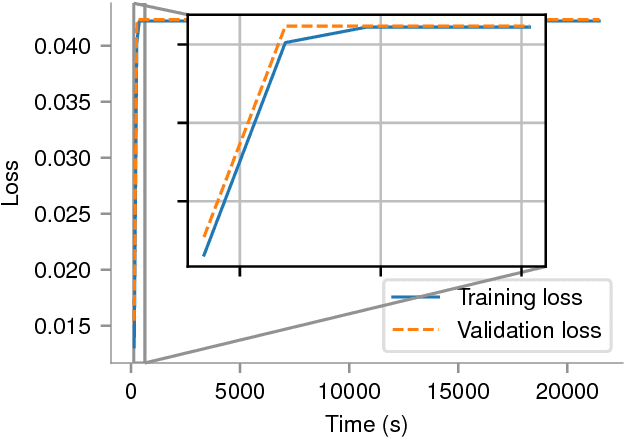
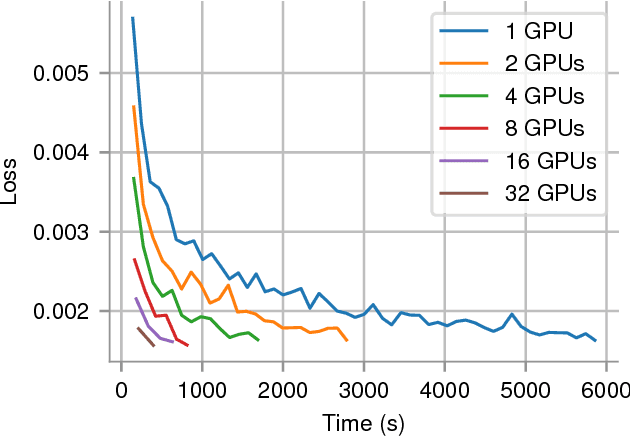
Abstract:In recent years, with the popularization of deep learning frameworks and large datasets, researchers have started parallelizing their models in order to train faster. This is crucially important, because they typically explore many hyperparameters in order to find the best ones for their applications. This process is time consuming and, consequently, speeding up training improves productivity. One approach to parallelize deep learning models followed by many researchers is based on weak scaling. The minibatches increase in size as new GPUs are added to the system. In addition, new learning rates schedules have been proposed to fix optimization issues that occur with large minibatch sizes. In this paper, however, we show that the recommendations provided by recent work do not apply to models that lack large datasets. In fact, we argument in favor of using strong scaling for achieving reliable performance in such cases. We evaluated our approach with up to 32 GPUs and show that weak scaling not only does not have the same accuracy as the sequential model, it also fails to converge most of time. Meanwhile, strong scaling has good scalability while having exactly the same accuracy of a sequential implementation.
Automatic tracking of vessel-like structures from a single starting point
Jun 08, 2017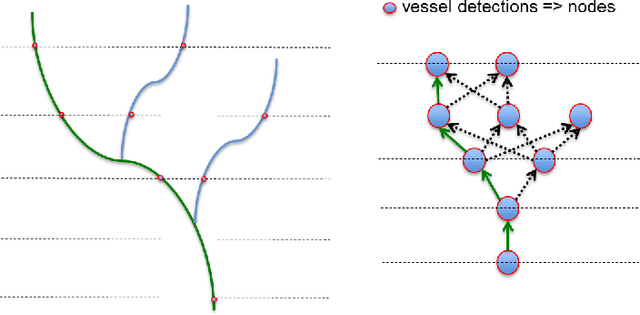

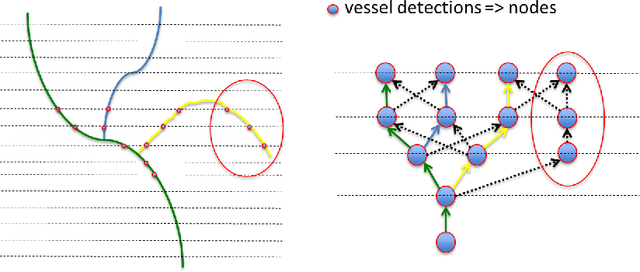
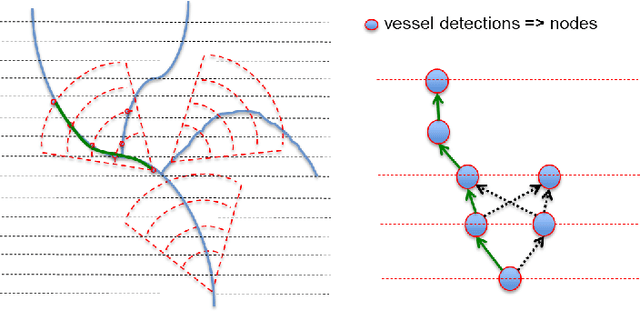
Abstract:The identification of vascular networks is an important topic in the medical image analysis community. While most methods focus on single vessel tracking, the few solutions that exist for tracking complete vascular networks are usually computationally intensive and require a lot of user interaction. In this paper we present a method to track full vascular networks iteratively using a single starting point. Our approach is based on a cloud of sampling points distributed over concentric spherical layers. We also proposed a vessel model and a metric of how well a sample point fits this model. Then, we implement the network tracking as a min-cost flow problem, and propose a novel optimization scheme to iteratively track the vessel structure by inherently handling bifurcations and paths. The method was tested using both synthetic and real images. On the 9 different data-sets of synthetic blood vessels, we achieved maximum accuracies of more than 98\%. We further use the synthetic data-set to analyse the sensibility of our method to parameter setting, showing the robustness of the proposed algorithm. For real images, we used coronary, carotid and pulmonary data to segment vascular structures and present the visual results. Still for real images, we present numerical and visual results for networks of nerve fibers in the olfactory system. Further visual results also show the potential of our approach for identifying vascular networks topologies. The presented method delivers good results for the several different datasets tested and have potential for segmenting vessel-like structures. Also, the topology information, inherently extracted, can be used for further analysis to computed aided diagnosis and surgical planning. Finally, the method's modular aspect holds potential for problem-oriented adjustments and improvements.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge