Daniel Racoceanu
LAB, IPAAL
ADNP-15: An Open-Source Histopathological Dataset for Neuritic Plaque Segmentation in Human Brain Whole Slide Images with Frequency Domain Image Enhancement for Stain Normalization
May 08, 2025Abstract:Alzheimer's Disease (AD) is a neurodegenerative disorder characterized by amyloid-beta plaques and tau neurofibrillary tangles, which serve as key histopathological features. The identification and segmentation of these lesions are crucial for understanding AD progression but remain challenging due to the lack of large-scale annotated datasets and the impact of staining variations on automated image analysis. Deep learning has emerged as a powerful tool for pathology image segmentation; however, model performance is significantly influenced by variations in staining characteristics, necessitating effective stain normalization and enhancement techniques. In this study, we address these challenges by introducing an open-source dataset (ADNP-15) of neuritic plaques (i.e., amyloid deposits combined with a crown of dystrophic tau-positive neurites) in human brain whole slide images. We establish a comprehensive benchmark by evaluating five widely adopted deep learning models across four stain normalization techniques, providing deeper insights into their influence on neuritic plaque segmentation. Additionally, we propose a novel image enhancement method that improves segmentation accuracy, particularly in complex tissue structures, by enhancing structural details and mitigating staining inconsistencies. Our experimental results demonstrate that this enhancement strategy significantly boosts model generalization and segmentation accuracy. All datasets and code are open-source, ensuring transparency and reproducibility while enabling further advancements in the field.
Performance Estimation for Supervised Medical Image Segmentation Models on Unlabeled Data Using UniverSeg
Apr 22, 2025Abstract:The performance of medical image segmentation models is usually evaluated using metrics like the Dice score and Hausdorff distance, which compare predicted masks to ground truth annotations. However, when applying the model to unseen data, such as in clinical settings, it is often impractical to annotate all the data, making the model's performance uncertain. To address this challenge, we propose the Segmentation Performance Evaluator (SPE), a framework for estimating segmentation models' performance on unlabeled data. This framework is adaptable to various evaluation metrics and model architectures. Experiments on six publicly available datasets across six evaluation metrics including pixel-based metrics such as Dice score and distance-based metrics like HD95, demonstrated the versatility and effectiveness of our approach, achieving a high correlation (0.956$\pm$0.046) and low MAE (0.025$\pm$0.019) compare with real Dice score on the independent test set. These results highlight its ability to reliably estimate model performance without requiring annotations. The SPE framework integrates seamlessly into any model training process without adding training overhead, enabling performance estimation and facilitating the real-world application of medical image segmentation algorithms. The source code is publicly available
Scalable, Trustworthy Generative Model for Virtual Multi-Staining from H&E Whole Slide Images
Jun 26, 2024



Abstract:Chemical staining methods are dependable but require extensive time, expensive chemicals, and raise environmental concerns. These challenges highlight the need for alternative solutions like virtual staining, which accelerates the diagnostic process and enhances stain application flexibility. Generative AI technologies are pivotal in addressing these issues. However, the high-stakes nature of healthcare decisions, especially in computational pathology, complicates the adoption of these tools due to their opaque processes. Our work introduces the use of generative AI for virtual staining, aiming to enhance performance, trustworthiness, scalability, and adaptability in computational pathology. The methodology centers on a singular H&E encoder supporting multiple stain decoders. This design focuses on critical regions in the latent space of H&E, enabling precise synthetic stain generation. Our method, tested to generate 8 different stains from a single H&E slide, offers scalability by loading only necessary model components during production. We integrate label-free knowledge in training, using loss functions and regularization to minimize artifacts, thus improving paired/unpaired virtual staining accuracy. To build trust, we use real-time self-inspection with discriminators for each stain type, providing pathologists with confidence heat-maps. Automatic quality checks on new H&E slides ensure conformity to the trained distribution, ensuring accurate synthetic stains. Recognizing pathologists' challenges with new technologies, we have developed an open-source, cloud-based system, that allows easy virtual staining of H&E slides through a browser, addressing hardware/software issues and facilitating real-time user feedback. We also curated a novel dataset of 8 paired H&E/stains related to pediatric Crohn's disease, comprising 480 WSIs to further stimulate computational pathology research.
Graph Theory and GNNs to Unravel the Topographical Organization of Brain Lesions in Variants of Alzheimer's Disease Progression
Mar 01, 2024



Abstract:This study utilizes graph theory and deep learning to assess variations in Alzheimer's disease (AD) neuropathologies, focusing on classic (cAD) and rapid (rpAD) progression forms. It analyses the distribution of amyloid plaques and tau tangles in postmortem brain tissues. Histopathological images are converted into tau-pathology-based graphs, and derived metrics are used for statistical analysis and in machine learning classifiers. These classifiers incorporate SHAP value explainability to differentiate between cAD and rpAD. Graph neural networks (GNNs) demonstrate greater efficiency than traditional CNN methods in analyzing this data, preserving spatial pathology context. Additionally, GNNs provide significant insights through explainable AI techniques. The analysis shows denser networks in rpAD and a distinctive impact on brain cortical layers: rpAD predominantly affects middle layers, whereas cAD influences both superficial and deep layers of the same cortical regions. These results suggest a unique neuropathological network organization for each AD variant.
Phagocytosis Unveiled: A Scalable and Interpretable Deep learning Framework for Neurodegenerative Disease Analysis
Apr 26, 2023Abstract:Quantifying the phagocytosis of dynamic, unstained cells is essential for evaluating neurodegenerative diseases. However, measuring rapid cell interactions and distinguishing cells from backgrounds make this task challenging when processing time-lapse phase-contrast video microscopy. In this study, we introduce a fully automated, scalable, and versatile realtime framework for quantifying and analyzing phagocytic activity. Our proposed pipeline can process large data-sets and includes a data quality verification module to counteract potential perturbations such as microscope movements and frame blurring. We also propose an explainable cell segmentation module to improve the interpretability of deep learning methods compared to black-box algorithms. This includes two interpretable deep learning capabilities: visual explanation and model simplification. We demonstrate that interpretability in deep learning is not the opposite of high performance, but rather provides essential deep learning algorithm optimization insights and solutions. Incorporating interpretable modules results in an efficient architecture design and optimized execution time. We apply this pipeline to quantify and analyze microglial cell phagocytosis in frontotemporal dementia (FTD) and obtain statistically reliable results showing that FTD mutant cells are larger and more aggressive than control cells. To stimulate translational approaches and future research, we release an open-source pipeline and a unique microglial cells phagocytosis dataset for immune system characterization in neurodegenerative diseases research. This pipeline and dataset will consistently crystallize future advances in this field, promoting the development of efficient and effective interpretable algorithms dedicated to this critical domain. https://github.com/ounissimehdi/PhagoStat
Frequency Disentangled Learning for Segmentation of Midbrain Structures from Quantitative Susceptibility Mapping Data
Feb 25, 2023



Abstract:One often lacks sufficient annotated samples for training deep segmentation models. This is in particular the case for less common imaging modalities such as Quantitative Susceptibility Mapping (QSM). It has been shown that deep models tend to fit the target function from low to high frequencies. One may hypothesize that such property can be leveraged for better training of deep learning models. In this paper, we exploit this property to propose a new training method based on frequency-domain disentanglement. It consists of two main steps: i) disentangling the image into high- and low-frequency parts and feature learning; ii) frequency-domain fusion to complete the task. The approach can be used with any backbone segmentation network. We apply the approach to the segmentation of the red and dentate nuclei from QSM data which is particularly relevant for the study of parkinsonian syndromes. We demonstrate that the proposed method provides considerable performance improvements for these tasks. We further applied it to three public datasets from the Medical Segmentation Decathlon (MSD) challenge. For two MSD tasks, it provided smaller but still substantial improvements (up to 7 points of Dice), especially under small training set situations.
Computational Pathology for Brain Disorders
Jan 13, 2023Abstract:Non-invasive brain imaging techniques allow understanding the behavior and macro changes in the brain to determine the progress of a disease. However, computational pathology provides a deeper understanding of brain disorders at cellular level, able to consolidate a diagnosis and make the bridge between the medical image and the omics analysis. In traditional histopathology, histology slides are visually inspected, under the microscope, by trained pathologists. This process is time-consuming and labor-intensive; therefore, the emergence of Computational Pathology has triggered great hope to ease this tedious task and make it more robust. This chapter focuses on understanding the state-of-the-art machine learning techniques used to analyze whole slide images within the context of brain disorders. We present a selective set of remarkable machine learning algorithms providing discriminative approaches and quality results on brain disorders. These methodologies are applied to different tasks, such as monitoring mechanisms contributing to disease progression and patient survival rates, analyzing morphological phenotypes for classification and quantitative assessment of disease, improving clinical care, diagnosing tumor specimens, and intraoperative interpretation. Thanks to the recent progress in machine learning algorithms for high-content image processing, computational pathology marks the rise of a new generation of medical discoveries and clinical protocols, including in brain disorders.
Fourier Disentangled Multimodal Prior Knowledge Fusion for Red Nucleus Segmentation in Brain MRI
Nov 02, 2022



Abstract:Early and accurate diagnosis of parkinsonian syndromes is critical to provide appropriate care to patients and for inclusion in therapeutic trials. The red nucleus is a structure of the midbrain that plays an important role in these disorders. It can be visualized using iron-sensitive magnetic resonance imaging (MRI) sequences. Different iron-sensitive contrasts can be produced with MRI. Combining such multimodal data has the potential to improve segmentation of the red nucleus. Current multimodal segmentation algorithms are computationally consuming, cannot deal with missing modalities and need annotations for all modalities. In this paper, we propose a new model that integrates prior knowledge from different contrasts for red nucleus segmentation. The method consists of three main stages. First, it disentangles the image into high-level information representing the brain structure, and low-frequency information representing the contrast. The high-frequency information is then fed into a network to learn anatomical features, while the list of multimodal low-frequency information is processed by another module. Finally, feature fusion is performed to complete the segmentation task. The proposed method was used with several iron-sensitive contrasts (iMag, QSM, R2*, SWI). Experiments demonstrate that our proposed model substantially outperforms a baseline UNet model when the training set size is very small.
eXclusive Autoencoder (XAE) for Nucleus Detection and Classification on Hematoxylin and Eosin (H&E) Stained Histopathological Images
Nov 27, 2018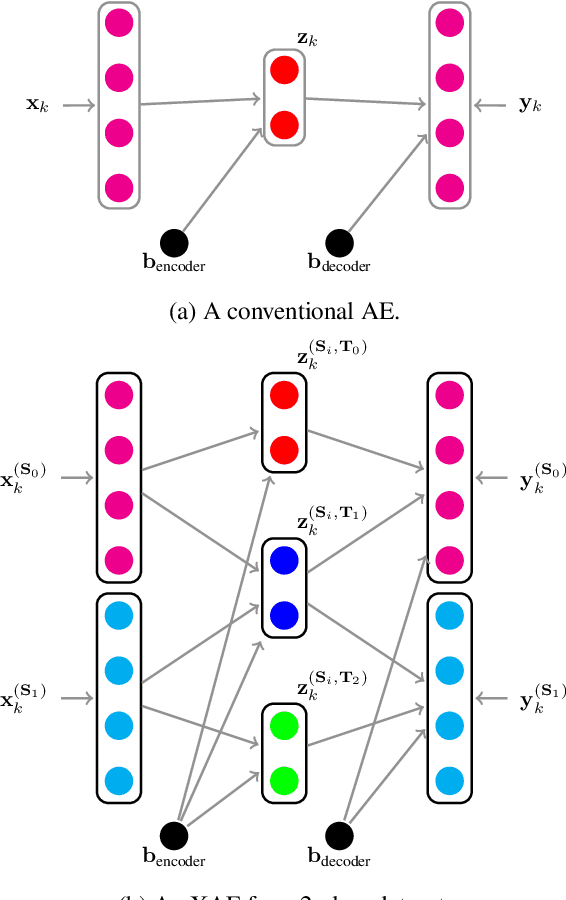
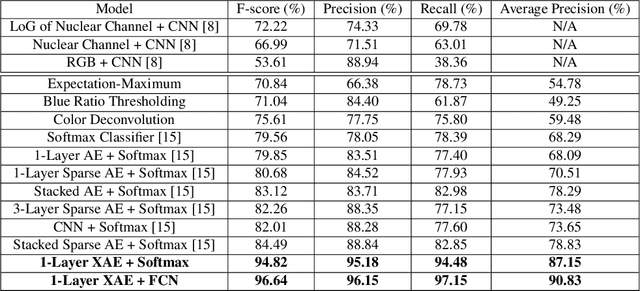
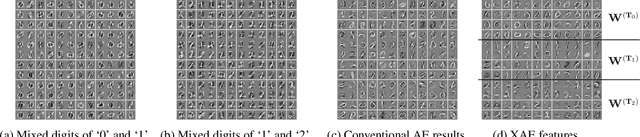
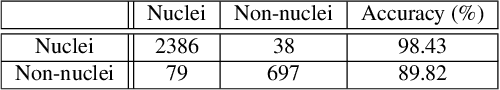
Abstract:In this paper, we introduced a novel feature extraction approach, named exclusive autoencoder (XAE), which is a supervised version of autoencoder (AE), able to largely improve the performance of nucleus detection and classification on hematoxylin and eosin (H&E) histopathological images. The proposed XAE can be used in any AE-based algorithm, as long as the data labels are also provided in the feature extraction phase. In the experiments, we evaluated the performance of an approach which is the combination of an XAE and a fully connected neural network (FCN) and compared with some AE-based methods. For a nucleus detection problem (considered as a nucleus/non-nucleus classification problem) on breast cancer H&E images, the F-score of the proposed XAE+FCN approach achieved 96.64% while the state-of-the-art was at 84.49%. For nucleus classification on colorectal cancer H&E images, with the annotations of four categories of epithelial, inflammatory, fibroblast and miscellaneous nuclei. The F-score of the proposed method reached 70.4%. We also proposed a lymphocyte segmentation method. In the step of lymphocyte detection, we have compared with cutting-edge technology and gained improved performance from 90% to 98.67%. We also proposed an algorithm for lymphocyte segmentation based on nucleus detection and classification. The obtained Dice coefficient achieved 88.31% while the cutting-edge approach was at 74%.
Gland Segmentation in Colon Histology Images: The GlaS Challenge Contest
Sep 01, 2016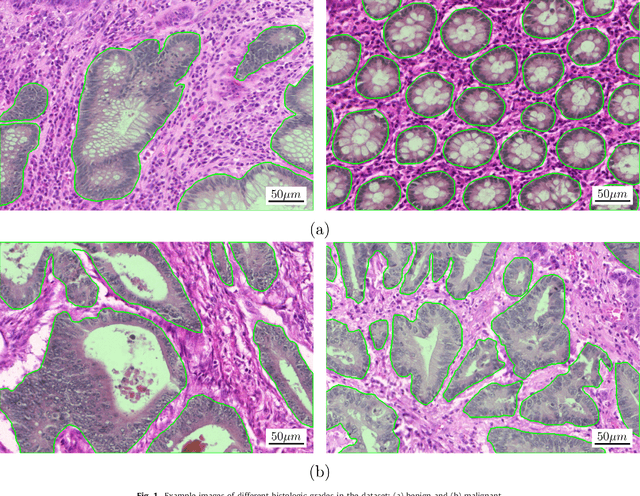
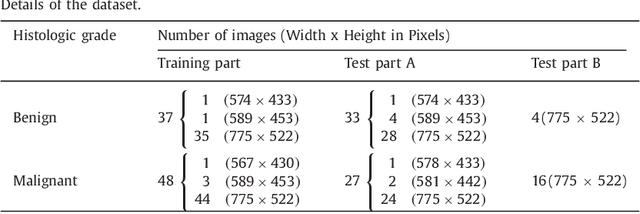
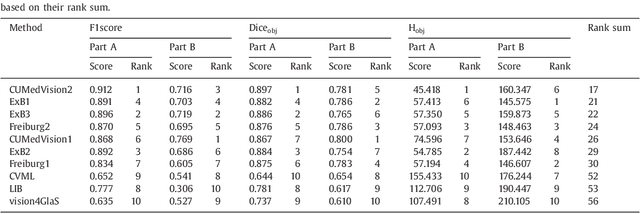
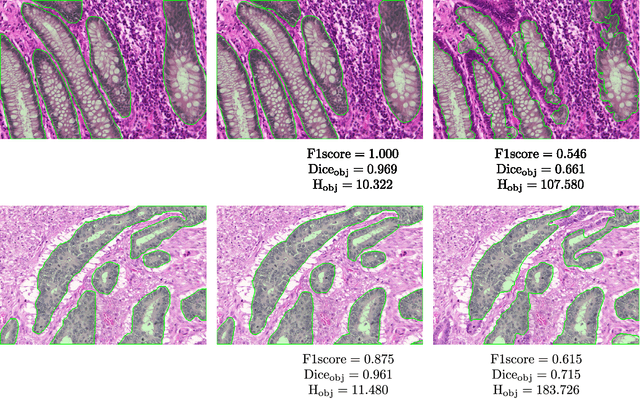
Abstract:Colorectal adenocarcinoma originating in intestinal glandular structures is the most common form of colon cancer. In clinical practice, the morphology of intestinal glands, including architectural appearance and glandular formation, is used by pathologists to inform prognosis and plan the treatment of individual patients. However, achieving good inter-observer as well as intra-observer reproducibility of cancer grading is still a major challenge in modern pathology. An automated approach which quantifies the morphology of glands is a solution to the problem. This paper provides an overview to the Gland Segmentation in Colon Histology Images Challenge Contest (GlaS) held at MICCAI'2015. Details of the challenge, including organization, dataset and evaluation criteria, are presented, along with the method descriptions and evaluation results from the top performing methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge