Lydia Chougar
Frequency Disentangled Learning for Segmentation of Midbrain Structures from Quantitative Susceptibility Mapping Data
Feb 25, 2023



Abstract:One often lacks sufficient annotated samples for training deep segmentation models. This is in particular the case for less common imaging modalities such as Quantitative Susceptibility Mapping (QSM). It has been shown that deep models tend to fit the target function from low to high frequencies. One may hypothesize that such property can be leveraged for better training of deep learning models. In this paper, we exploit this property to propose a new training method based on frequency-domain disentanglement. It consists of two main steps: i) disentangling the image into high- and low-frequency parts and feature learning; ii) frequency-domain fusion to complete the task. The approach can be used with any backbone segmentation network. We apply the approach to the segmentation of the red and dentate nuclei from QSM data which is particularly relevant for the study of parkinsonian syndromes. We demonstrate that the proposed method provides considerable performance improvements for these tasks. We further applied it to three public datasets from the Medical Segmentation Decathlon (MSD) challenge. For two MSD tasks, it provided smaller but still substantial improvements (up to 7 points of Dice), especially under small training set situations.
Fourier Disentangled Multimodal Prior Knowledge Fusion for Red Nucleus Segmentation in Brain MRI
Nov 02, 2022



Abstract:Early and accurate diagnosis of parkinsonian syndromes is critical to provide appropriate care to patients and for inclusion in therapeutic trials. The red nucleus is a structure of the midbrain that plays an important role in these disorders. It can be visualized using iron-sensitive magnetic resonance imaging (MRI) sequences. Different iron-sensitive contrasts can be produced with MRI. Combining such multimodal data has the potential to improve segmentation of the red nucleus. Current multimodal segmentation algorithms are computationally consuming, cannot deal with missing modalities and need annotations for all modalities. In this paper, we propose a new model that integrates prior knowledge from different contrasts for red nucleus segmentation. The method consists of three main stages. First, it disentangles the image into high-level information representing the brain structure, and low-frequency information representing the contrast. The high-frequency information is then fed into a network to learn anatomical features, while the list of multimodal low-frequency information is processed by another module. Finally, feature fusion is performed to complete the segmentation task. The proposed method was used with several iron-sensitive contrasts (iMag, QSM, R2*, SWI). Experiments demonstrate that our proposed model substantially outperforms a baseline UNet model when the training set size is very small.
Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks
Sep 11, 2018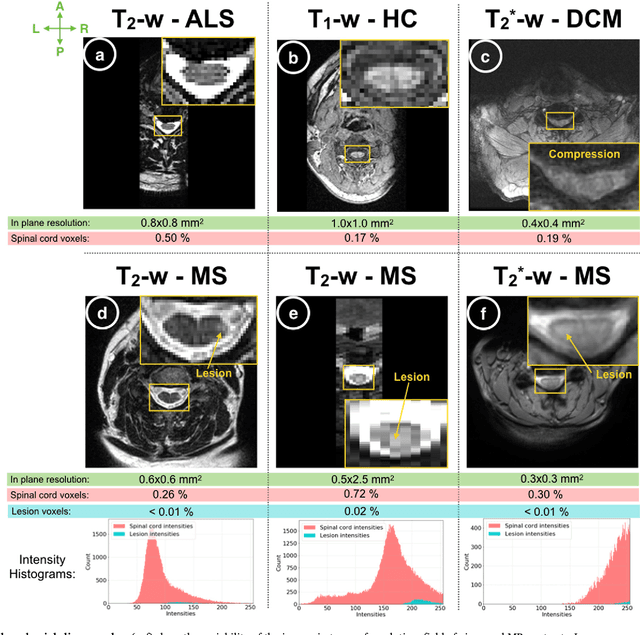
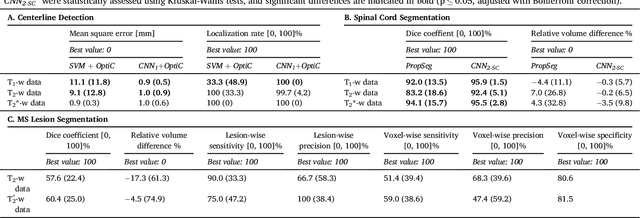
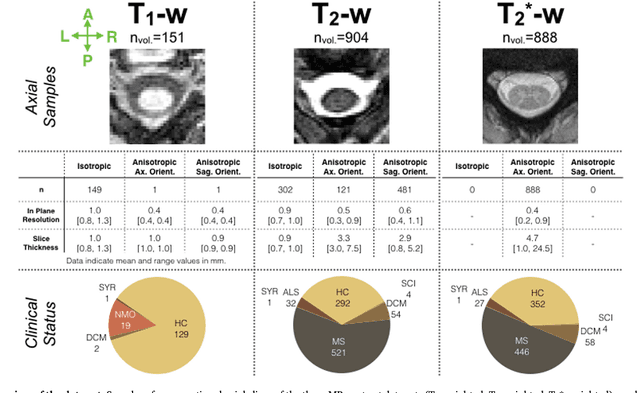
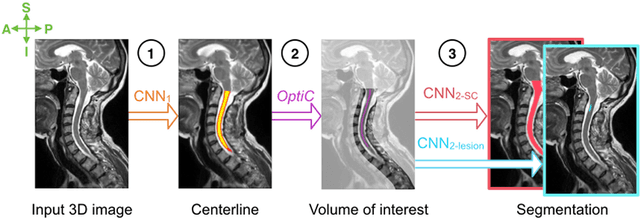
Abstract:The spinal cord is frequently affected by atrophy and/or lesions in multiple sclerosis (MS) patients. Segmentation of the spinal cord and lesions from MRI data provides measures of damage, which are key criteria for the diagnosis, prognosis, and longitudinal monitoring in MS. Automating this operation eliminates inter-rater variability and increases the efficiency of large-throughput analysis pipelines. Robust and reliable segmentation across multi-site spinal cord data is challenging because of the large variability related to acquisition parameters and image artifacts. The goal of this study was to develop a fully-automatic framework, robust to variability in both image parameters and clinical condition, for segmentation of the spinal cord and intramedullary MS lesions from conventional MRI data. Scans of 1,042 subjects (459 healthy controls, 471 MS patients, and 112 with other spinal pathologies) were included in this multi-site study (n=30). Data spanned three contrasts (T1-, T2-, and T2*-weighted) for a total of 1,943 volumes. The proposed cord and lesion automatic segmentation approach is based on a sequence of two Convolutional Neural Networks (CNNs). To deal with the very small proportion of spinal cord and/or lesion voxels compared to the rest of the volume, a first CNN with 2D dilated convolutions detects the spinal cord centerline, followed by a second CNN with 3D convolutions that segments the spinal cord and/or lesions. When compared against manual segmentation, our CNN-based approach showed a median Dice of 95% vs. 88% for PropSeg, a state-of-the-art spinal cord segmentation method. Regarding lesion segmentation on MS data, our framework provided a Dice of 60%, a relative volume difference of -15%, and a lesion-wise detection sensitivity and precision of 83% and 77%, respectively. The proposed framework is open-source and readily available in the Spinal Cord Toolbox.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge