Daniel Tse
The Llama 4 Herd: Architecture, Training, Evaluation, and Deployment Notes
Jan 15, 2026Abstract:This document consolidates publicly reported technical details about Metas Llama 4 model family. It summarizes (i) released variants (Scout and Maverick) and the broader herd context including the previewed Behemoth teacher model, (ii) architectural characteristics beyond a high-level MoE description covering routed/shared-expert structure, early-fusion multimodality, and long-context design elements reported for Scout (iRoPE and length generalization strategies), (iii) training disclosures spanning pre-training, mid-training for long-context extension, and post-training methodology (lightweight SFT, online RL, and lightweight DPO) as described in release materials, (iv) developer-reported benchmark results for both base and instruction-tuned checkpoints, and (v) practical deployment constraints observed across major serving environments, including provider-specific context limits and quantization packaging. The manuscript also summarizes licensing obligations relevant to redistribution and derivative naming, and reviews publicly described safeguards and evaluation practices. The goal is to provide a compact technical reference for researchers and practitioners who need precise, source-backed facts about Llama 4.
Advancing Multimodal Medical Capabilities of Gemini
May 06, 2024



Abstract:Many clinical tasks require an understanding of specialized data, such as medical images and genomics, which is not typically found in general-purpose large multimodal models. Building upon Gemini's multimodal models, we develop several models within the new Med-Gemini family that inherit core capabilities of Gemini and are optimized for medical use via fine-tuning with 2D and 3D radiology, histopathology, ophthalmology, dermatology and genomic data. Med-Gemini-2D sets a new standard for AI-based chest X-ray (CXR) report generation based on expert evaluation, exceeding previous best results across two separate datasets by an absolute margin of 1% and 12%, where 57% and 96% of AI reports on normal cases, and 43% and 65% on abnormal cases, are evaluated as "equivalent or better" than the original radiologists' reports. We demonstrate the first ever large multimodal model-based report generation for 3D computed tomography (CT) volumes using Med-Gemini-3D, with 53% of AI reports considered clinically acceptable, although additional research is needed to meet expert radiologist reporting quality. Beyond report generation, Med-Gemini-2D surpasses the previous best performance in CXR visual question answering (VQA) and performs well in CXR classification and radiology VQA, exceeding SoTA or baselines on 17 of 20 tasks. In histopathology, ophthalmology, and dermatology image classification, Med-Gemini-2D surpasses baselines across 18 out of 20 tasks and approaches task-specific model performance. Beyond imaging, Med-Gemini-Polygenic outperforms the standard linear polygenic risk score-based approach for disease risk prediction and generalizes to genetically correlated diseases for which it has never been trained. Although further development and evaluation are necessary in the safety-critical medical domain, our results highlight the potential of Med-Gemini across a wide range of medical tasks.
ELIXR: Towards a general purpose X-ray artificial intelligence system through alignment of large language models and radiology vision encoders
Aug 02, 2023Abstract:Our approach, which we call Embeddings for Language/Image-aligned X-Rays, or ELIXR, leverages a language-aligned image encoder combined or grafted onto a fixed LLM, PaLM 2, to perform a broad range of tasks. We train this lightweight adapter architecture using images paired with corresponding free-text radiology reports from the MIMIC-CXR dataset. ELIXR achieved state-of-the-art performance on zero-shot chest X-ray (CXR) classification (mean AUC of 0.850 across 13 findings), data-efficient CXR classification (mean AUCs of 0.893 and 0.898 across five findings (atelectasis, cardiomegaly, consolidation, pleural effusion, and pulmonary edema) for 1% (~2,200 images) and 10% (~22,000 images) training data), and semantic search (0.76 normalized discounted cumulative gain (NDCG) across nineteen queries, including perfect retrieval on twelve of them). Compared to existing data-efficient methods including supervised contrastive learning (SupCon), ELIXR required two orders of magnitude less data to reach similar performance. ELIXR also showed promise on CXR vision-language tasks, demonstrating overall accuracies of 58.7% and 62.5% on visual question answering and report quality assurance tasks, respectively. These results suggest that ELIXR is a robust and versatile approach to CXR AI.
Enabling faster and more reliable sonographic assessment of gestational age through machine learning
Mar 22, 2022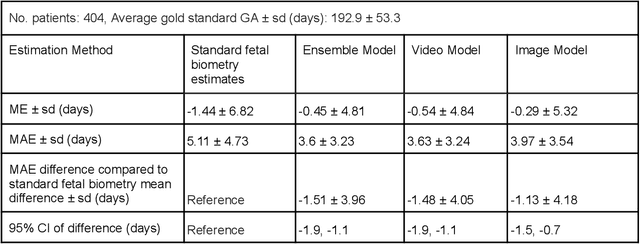
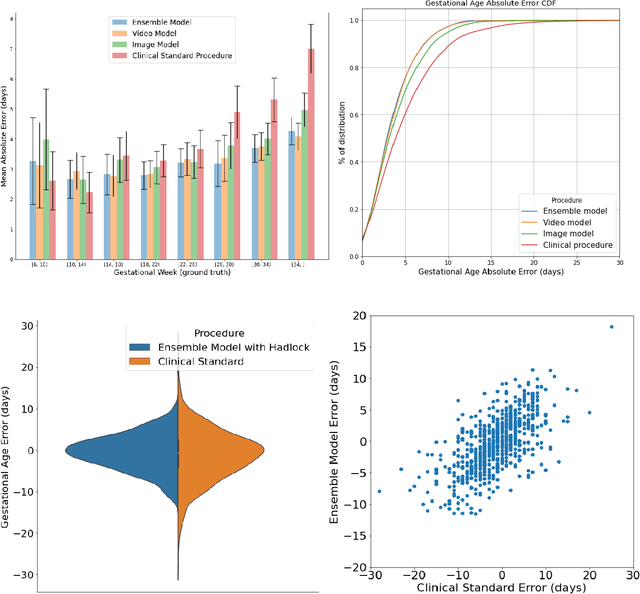
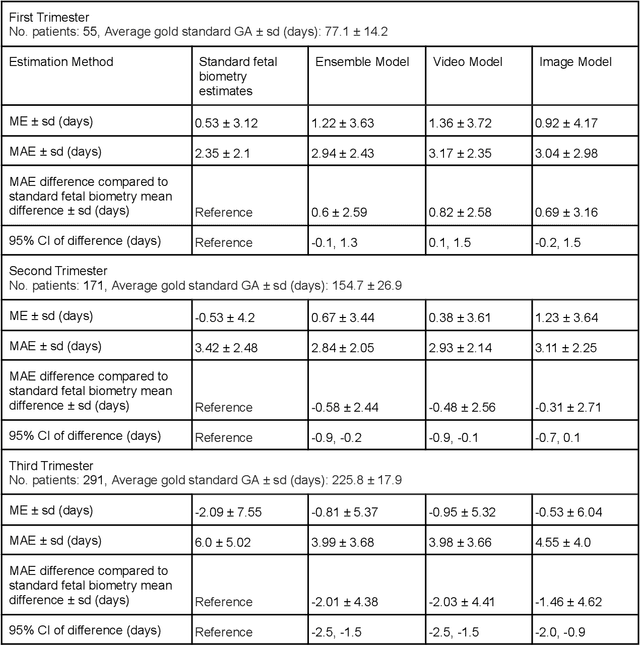
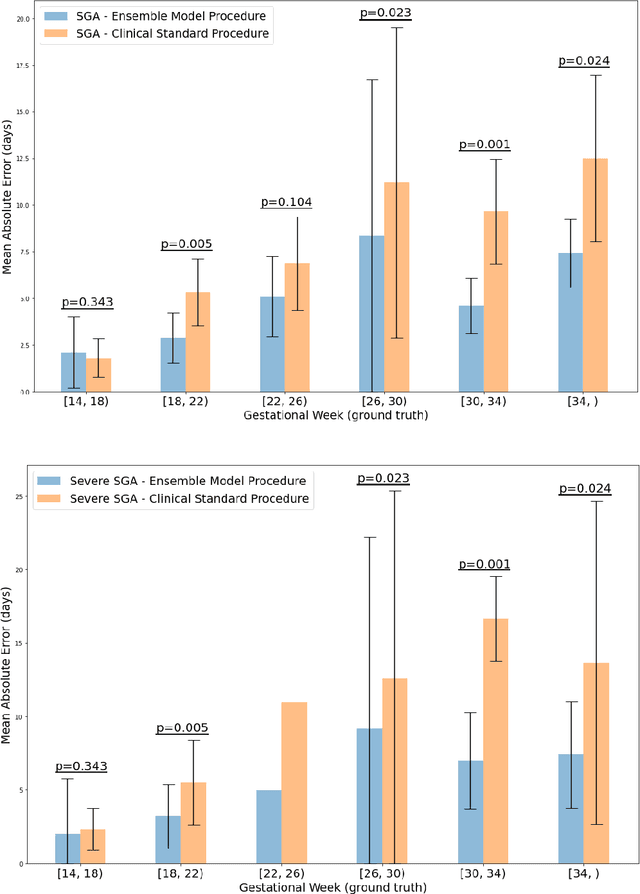
Abstract:Fetal ultrasounds are an essential part of prenatal care and can be used to estimate gestational age (GA). Accurate GA assessment is important for providing appropriate prenatal care throughout pregnancy and identifying complications such as fetal growth disorders. Since derivation of GA from manual fetal biometry measurements (head, abdomen, femur) are operator-dependent and time-consuming, there have been a number of research efforts focused on using artificial intelligence (AI) models to estimate GA using standard biometry images, but there is still room to improve the accuracy and reliability of these AI systems for widescale adoption. To improve GA estimates, without significant change to provider workflows, we leverage AI to interpret standard plane ultrasound images as well as 'fly-to' ultrasound videos, which are 5-10s videos automatically recorded as part of the standard of care before the still image is captured. We developed and validated three AI models: an image model using standard plane images, a video model using fly-to videos, and an ensemble model (combining both image and video). All three were statistically superior to standard fetal biometry-based GA estimates derived by expert sonographers, the ensemble model has the lowest mean absolute error (MAE) compared to the clinical standard fetal biometry (mean difference: -1.51 $\pm$ 3.96 days, 95% CI [-1.9, -1.1]) on a test set that consisted of 404 participants. We showed that our models outperform standard biometry by a more substantial margin on fetuses that were small for GA. Our AI models have the potential to empower trained operators to estimate GA with higher accuracy while reducing the amount of time required and user variability in measurement acquisition.
AI system for fetal ultrasound in low-resource settings
Mar 18, 2022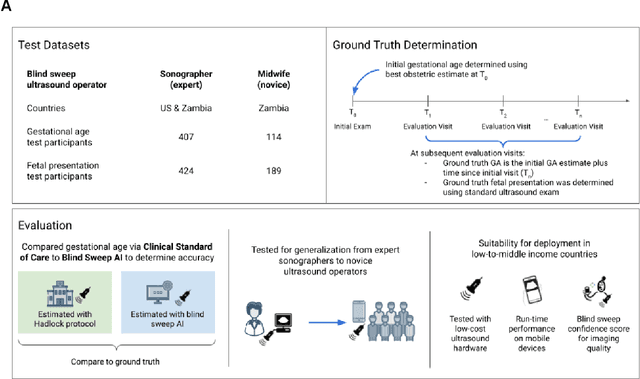
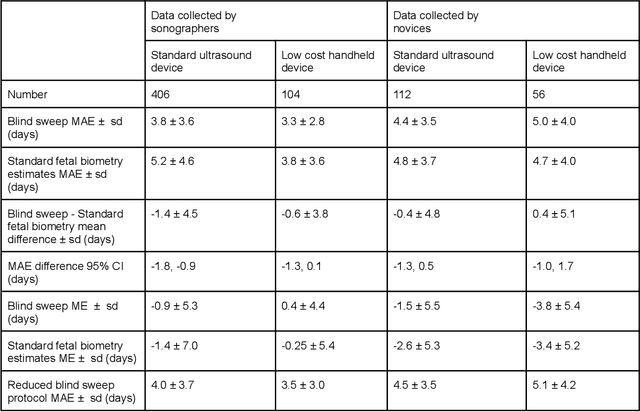
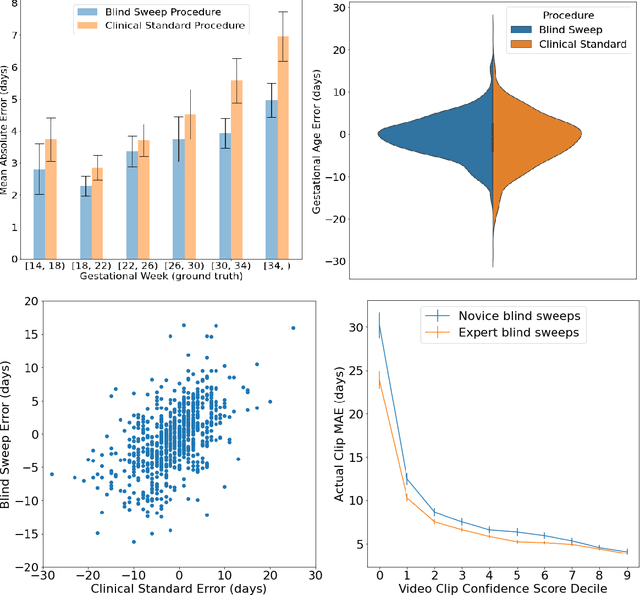
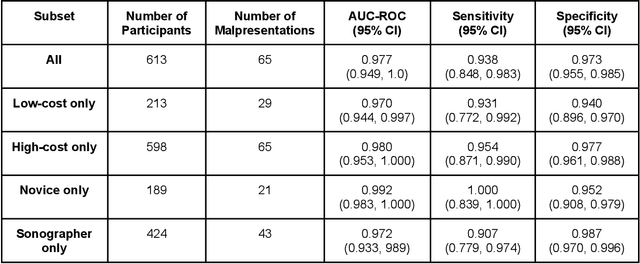
Abstract:Despite considerable progress in maternal healthcare, maternal and perinatal deaths remain high in low-to-middle income countries. Fetal ultrasound is an important component of antenatal care, but shortage of adequately trained healthcare workers has limited its adoption. We developed and validated an artificial intelligence (AI) system that uses novice-acquired "blind sweep" ultrasound videos to estimate gestational age (GA) and fetal malpresentation. We further addressed obstacles that may be encountered in low-resourced settings. Using a simplified sweep protocol with real-time AI feedback on sweep quality, we have demonstrated the generalization of model performance to minimally trained novice ultrasound operators using low cost ultrasound devices with on-device AI integration. The GA model was non-inferior to standard fetal biometry estimates with as few as two sweeps, and the fetal malpresentation model had high AUC-ROCs across operators and devices. Our AI models have the potential to assist in upleveling the capabilities of lightly trained ultrasound operators in low resource settings.
Deep learning for detecting pulmonary tuberculosis via chest radiography: an international study across 10 countries
May 16, 2021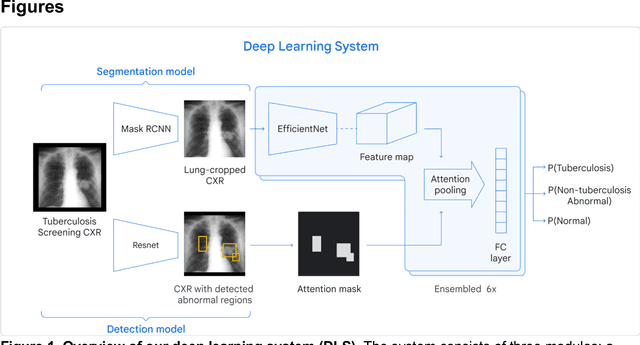
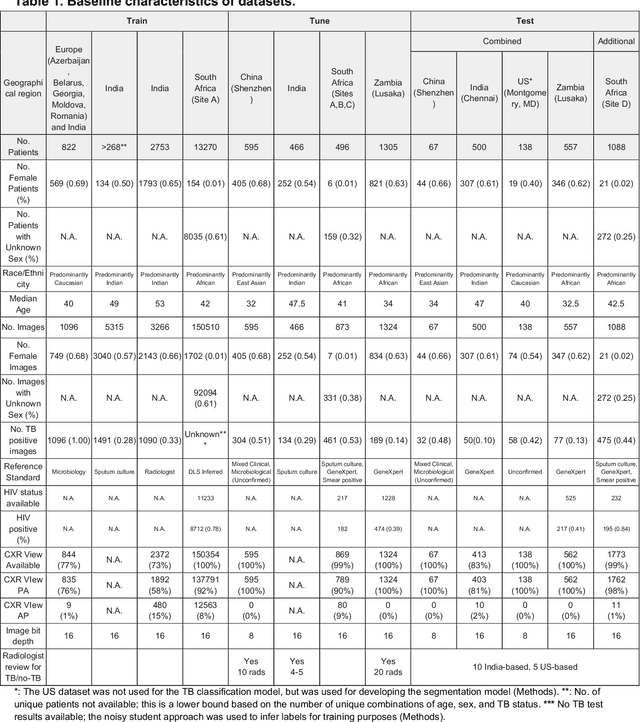
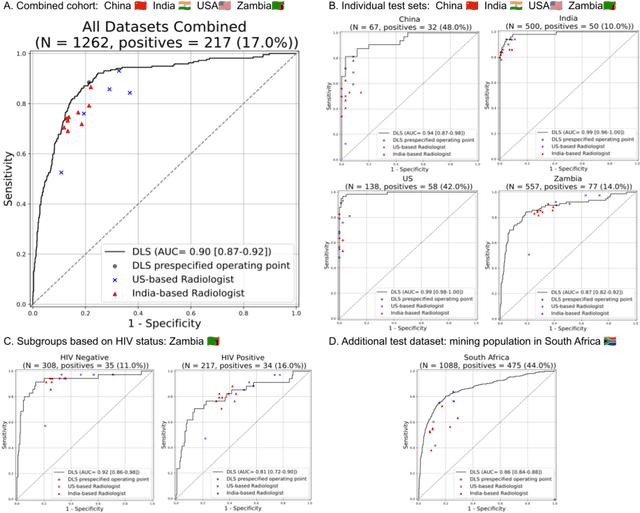
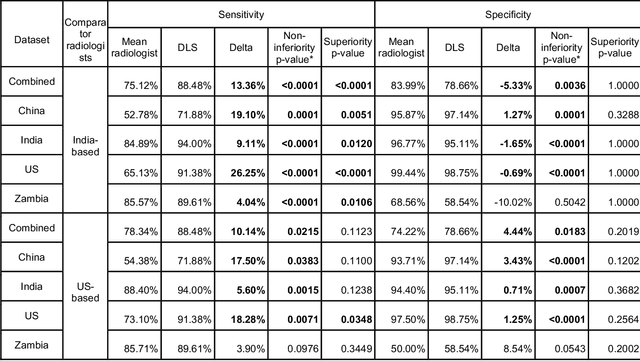
Abstract:Tuberculosis (TB) is a top-10 cause of death worldwide. Though the WHO recommends chest radiographs (CXRs) for TB screening, the limited availability of CXR interpretation is a barrier. We trained a deep learning system (DLS) to detect active pulmonary TB using CXRs from 9 countries across Africa, Asia, and Europe, and utilized large-scale CXR pretraining, attention pooling, and noisy student semi-supervised learning. Evaluation was on (1) a combined test set spanning China, India, US, and Zambia, and (2) an independent mining population in South Africa. Given WHO targets of 90% sensitivity and 70% specificity, the DLS's operating point was prespecified to favor sensitivity over specificity. On the combined test set, the DLS's ROC curve was above all 9 India-based radiologists, with an AUC of 0.90 (95%CI 0.87-0.92). The DLS's sensitivity (88%) was higher than the India-based radiologists (75% mean sensitivity), p<0.001 for superiority; and its specificity (79%) was non-inferior to the radiologists (84% mean specificity), p=0.004. Similar trends were observed within HIV positive and sputum smear positive sub-groups, and in the South Africa test set. We found that 5 US-based radiologists (where TB isn't endemic) were more sensitive and less specific than the India-based radiologists (where TB is endemic). The DLS also remained non-inferior to the US-based radiologists. In simulations, using the DLS as a prioritization tool for confirmatory testing reduced the cost per positive case detected by 40-80% compared to using confirmatory testing alone. To conclude, our DLS generalized to 5 countries, and merits prospective evaluation to assist cost-effective screening efforts in radiologist-limited settings. Operating point flexibility may permit customization of the DLS to account for site-specific factors such as TB prevalence, demographics, clinical resources, and customary practice patterns.
Interpretable Survival Prediction for Colorectal Cancer using Deep Learning
Nov 17, 2020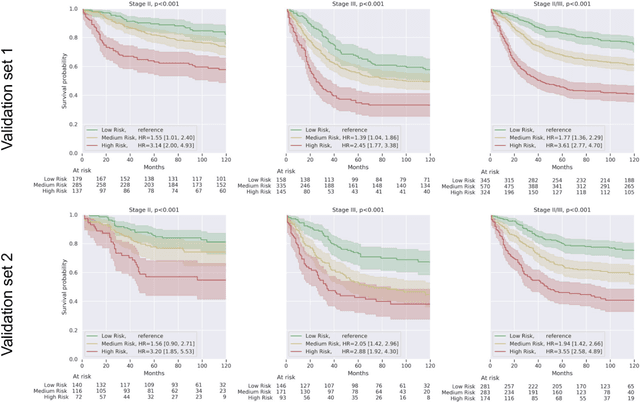
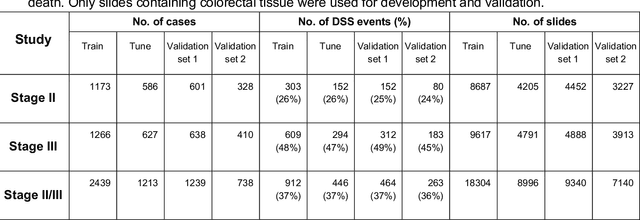
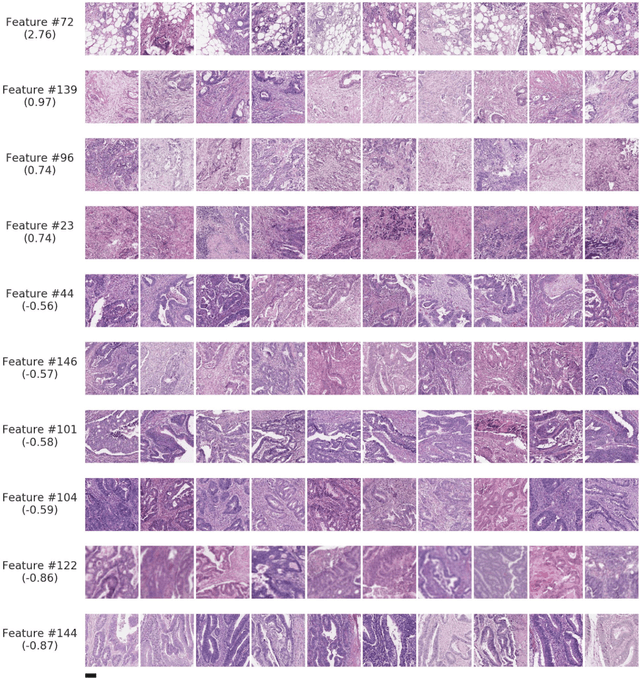
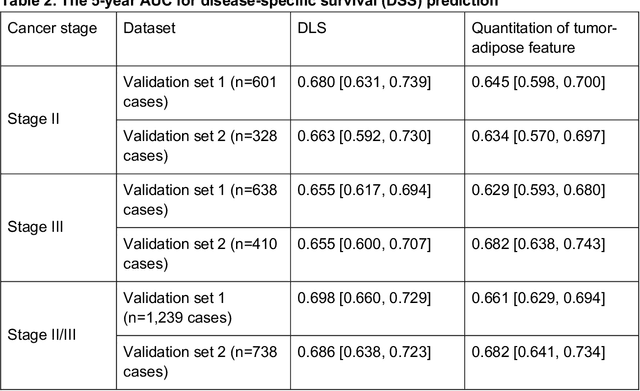
Abstract:Deriving interpretable prognostic features from deep-learning-based prognostic histopathology models remains a challenge. In this study, we developed a deep learning system (DLS) for predicting disease specific survival for stage II and III colorectal cancer using 3,652 cases (27,300 slides). When evaluated on two validation datasets containing 1,239 cases (9,340 slides) and 738 cases (7,140 slides) respectively, the DLS achieved a 5-year disease-specific survival AUC of 0.70 (95%CI 0.66-0.73) and 0.69 (95%CI 0.64-0.72), and added significant predictive value to a set of 9 clinicopathologic features. To interpret the DLS, we explored the ability of different human-interpretable features to explain the variance in DLS scores. We observed that clinicopathologic features such as T-category, N-category, and grade explained a small fraction of the variance in DLS scores (R2=18% in both validation sets). Next, we generated human-interpretable histologic features by clustering embeddings from a deep-learning based image-similarity model and showed that they explain the majority of the variance (R2 of 73% to 80%). Furthermore, the clustering-derived feature most strongly associated with high DLS scores was also highly prognostic in isolation. With a distinct visual appearance (poorly differentiated tumor cell clusters adjacent to adipose tissue), this feature was identified by annotators with 87.0-95.5% accuracy. Our approach can be used to explain predictions from a prognostic deep learning model and uncover potentially-novel prognostic features that can be reliably identified by people for future validation studies.
Deep Learning for Distinguishing Normal versus Abnormal Chest Radiographs and Generalization to Unseen Diseases
Oct 22, 2020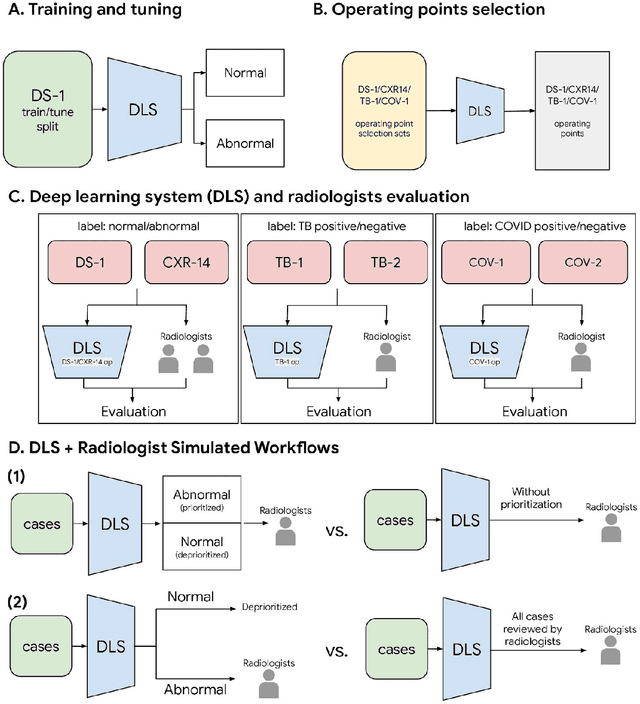
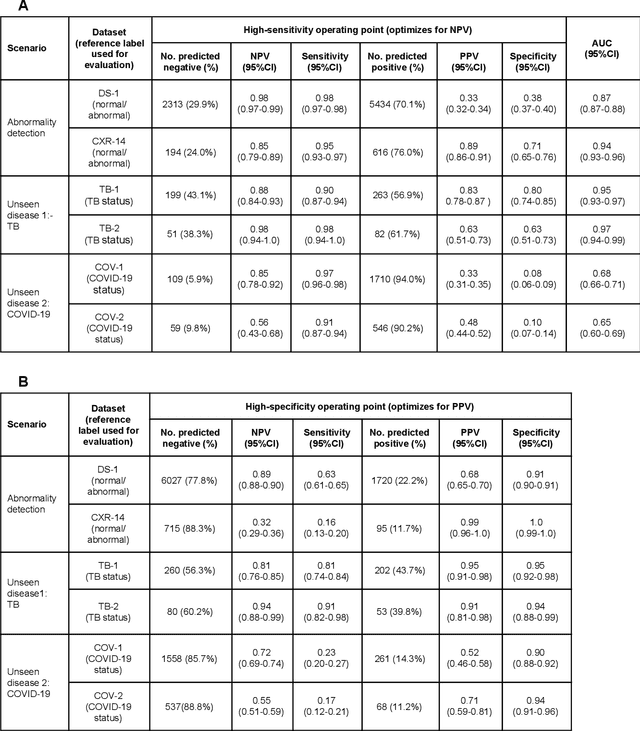
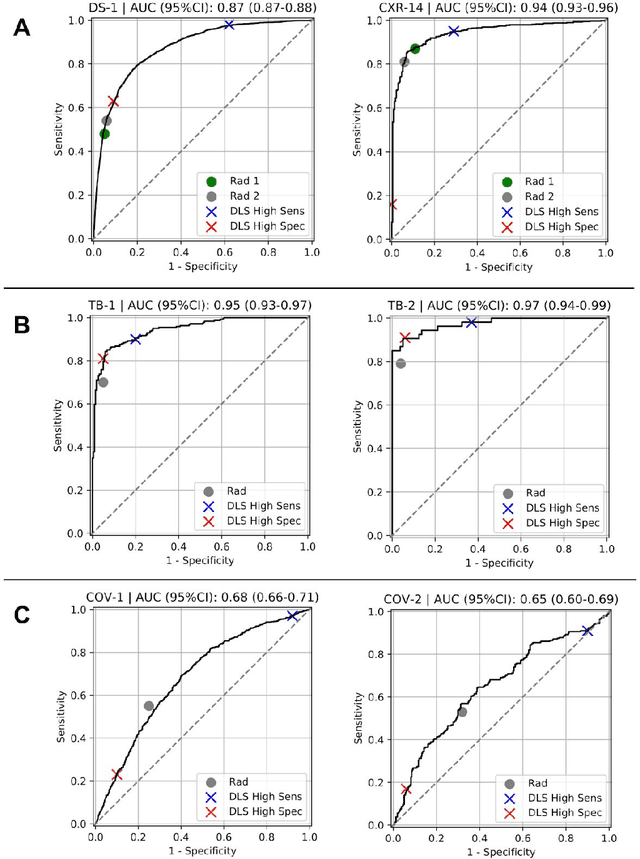
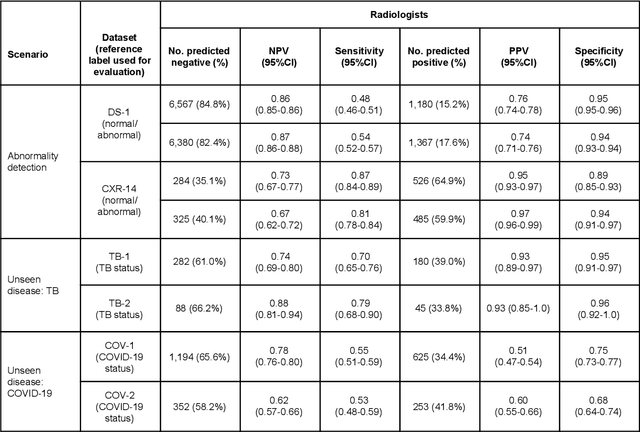
Abstract:Chest radiography (CXR) is the most widely-used thoracic clinical imaging modality and is crucial for guiding the management of cardiothoracic conditions. The detection of specific CXR findings has been the main focus of several artificial intelligence (AI) systems. However, the wide range of possible CXR abnormalities makes it impractical to build specific systems to detect every possible condition. In this work, we developed and evaluated an AI system to classify CXRs as normal or abnormal. For development, we used a de-identified dataset of 248,445 patients from a multi-city hospital network in India. To assess generalizability, we evaluated our system using 6 international datasets from India, China, and the United States. Of these datasets, 4 focused on diseases that the AI was not trained to detect: 2 datasets with tuberculosis and 2 datasets with coronavirus disease 2019. Our results suggest that the AI system generalizes to new patient populations and abnormalities. In a simulated workflow where the AI system prioritized abnormal cases, the turnaround time for abnormal cases reduced by 7-28%. These results represent an important step towards evaluating whether AI can be safely used to flag cases in a general setting where previously unseen abnormalities exist.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge